Fig. 5.
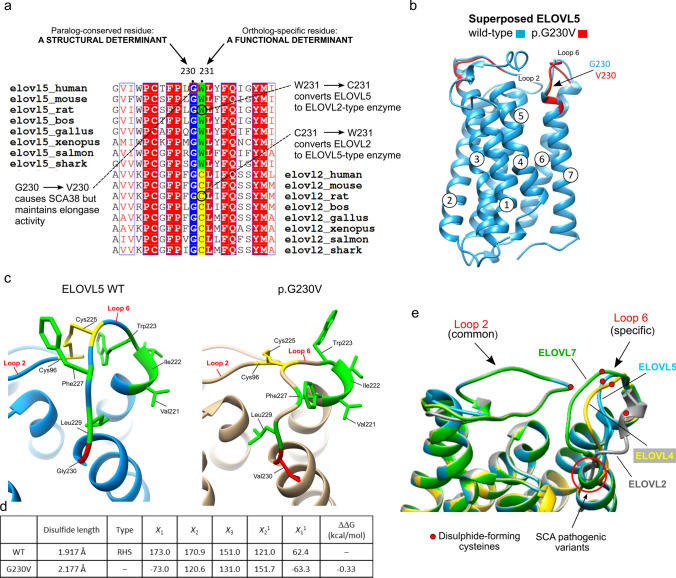
Evolutionary conservation of G230 in ELOVL5 and ELOVL2, and impact of p.G230V on protein conformation. a Relevant section of the multiple sequence alignment (MSA) of vertebrate ELOVL5 orthologs and its closest paralog ELOVL2. The full MSA is available in the Supporting Information. The MSA highlights that G230 is common to the paralogs ELOVL5 and ELOVL2, suggesting its relevance for structural integrity; the adjacent residue (ELOVL5 W231; ELOVL2 C217) is ortholog-specific, pointing to a functional role specific to a particular ELOVL protein. This is supported by the fact that changing Elovl5 W231 (in rat) to C231 modifies the substrate preference of ELOVL5 to that of ELOVL2, and vice versa (Gregory et al. 2013). b Overlay of the 3D structures of wild-type (WT) ELOVL5 (blue) and p.G230V (red) proteins highlighting the position of G230V, the correct superposition of Loop 2 which contrasts with the shift in position of Loop 6. c Enlargement of Loop 2/Loop 6 intramolecular bond, showing how the disulphide bond changes in p.G230V (in gold, on right) with respect to the bond in the wild-type protein (WT, blue, on left). Hydrophobic residues of Loop 6 are shown in green to highlight the positional change of their side chains. The conserved cysteines that form the disulphide bond are shown in yellow. d Table illustrates the impact of G230V substitution on the length and form of the conserved disulphide bond, and on protein stability. The length of the bonds were calculated and visualized in UCSF Chimera. The PDB files of wild-type and p.G230V were uploaded in the PDBsum home page (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/) to generate protein structural details including disulphide bond conformation. The program calculated the conformation of the conserved disulphide bond in wild-type and p.G230V ELOVL5, measuring the five dihedral angles (X1, X2, X3, X21 and X22) that form this bond. The conformation favoured by wild-type ELOVL5 is the right-handed spiral (RHS), which is lost in p.G230V. Negative free energy variation indicates a loss of protein stability, the result shown here was calculated with SAAFEC-SEQ. e The protein structures of native ELOVL5 (blue), ELOVL7 (green), ELOVL4 (yellow) and ELOVL2 (grey) were superimposed and zoomed to highlight the shared conformation of Loop 2 in the four elongases, in contrast to the individual ELOVL-specific conformation of Loop 6. The red dots represent the position of the conserved Cys residues that form the intramolecular disulphide bond. The position of the pathogenic variants ELOVL5 G230V and ELOVL4 W246G are shown (red arrow)