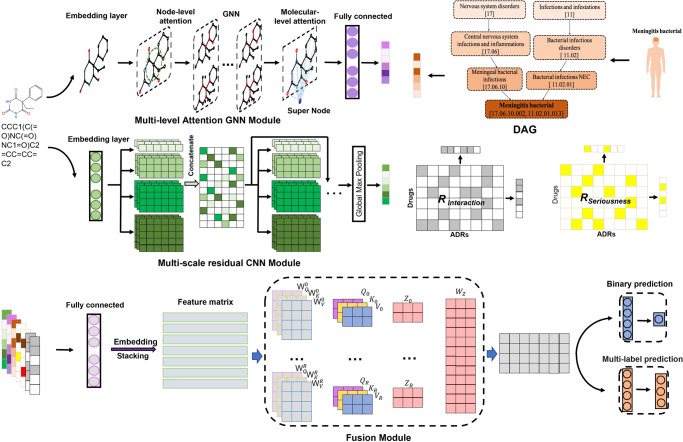
Fig. 2. The network architecture of GCAP.
Given a SMILES sequence of a drug and the semantic descriptors of an ADR, a drug molecule graph, a drug SMILES encoding matrix, and a semantic feature vector of the ADR can be constructed. The multi-level graph attention module and multi-scale residual CNN module are then used to extract representations from the drug molecule graph and SMILES encoding matrix, respectively. The semantic feature vector of an ADR is calculated based on all associated descriptors, and a fully connected layer is used to extract the representation from the semantic feature vector. Simultaneously, multiple fully-connected layers generate drug and ADR representations from the clinical outcomes of known drug–ADR interactions. All representations are then stacked and fed into a multi-head self-attention module to fuse multiple representations into a joint vector for downstream predictions. By setting different downstream classifiers for each task, GCAP can accurately predict potential drug–ADR interactions that cause serious clinical outcomes and identify the corresponding classes of serious clinical outcomes.