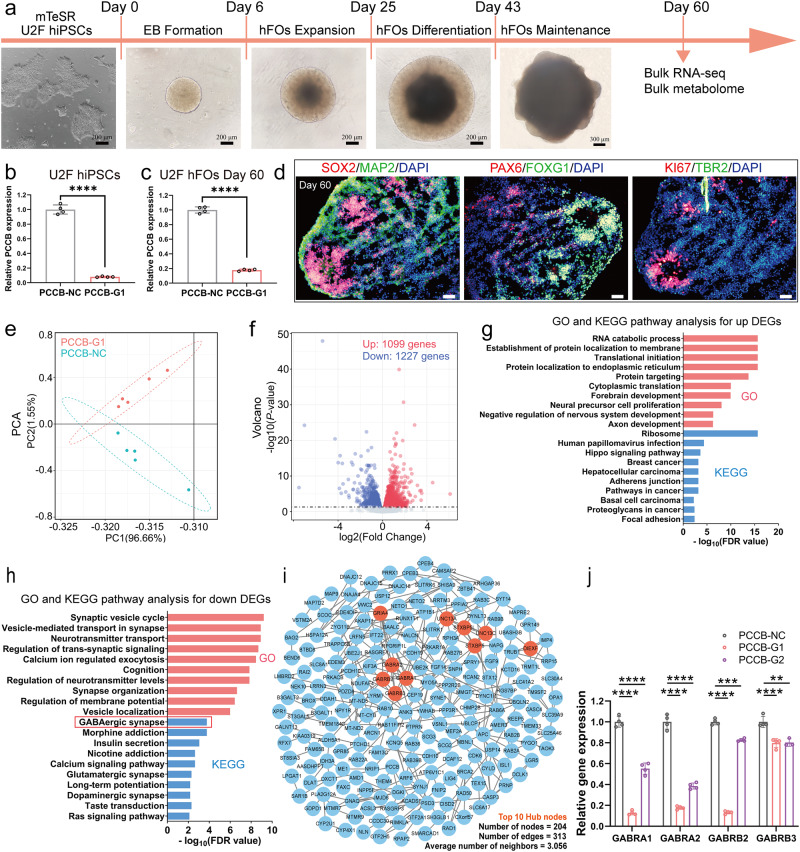
Fig. 2. Functional effects of PCCB knockdown in U2F hFOs.
a Workflow of hFOs culture in this study. Scale bar, 200 μm and 300 μm. b, c RT-qPCR analysis for PCCB expression in hiPSCs (b) and hFOs (c), n = four technical replicates per group. d Immunostaining characterization for representative control hFOs. N ≥ three biologically independent samples in each group. Cortical plate marker, MAP2; intermediate zone marker, TBR2; ventricular zone markers, SOX2 and ki67; forebrain-specific markers, FOXG1 and PAX6. Scale bar, 50 μm. e PCA plot for the PCCB knockdown and control hFOs. f Volcano plot of DEGs between the PCCB knockdown and control hFOs. Upregulated genes are shown with red dots and downregulated genes are shown with blue dots. g, h GO and KEGG analysis for the PCCB knockdown-induced upregulated (g) and downregulated DEGs (h), respectively. GO terms are shown with red bars, and KEGG pathways are shown with blue bars. i PPI network analysis for 350 downregulated DEGs shared in both PCCB-G1 and PCCB-G2 hFOs. The top 10 hub nodes are shown with orange nodes. j RT-qPCR analysis for GABRA1, GABRA2, GABRB2, and GABRB3 (The hub nodes in the PPI network), n = four technical replicates per group. Data are shown as Mean ± SD. The unpaired two-tailed t-test was used to assess difference between the PCCB-NC and PCCB-G1 or PCCB-G2 group. **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data underlying b, c, and j are provided as a Source Data file.