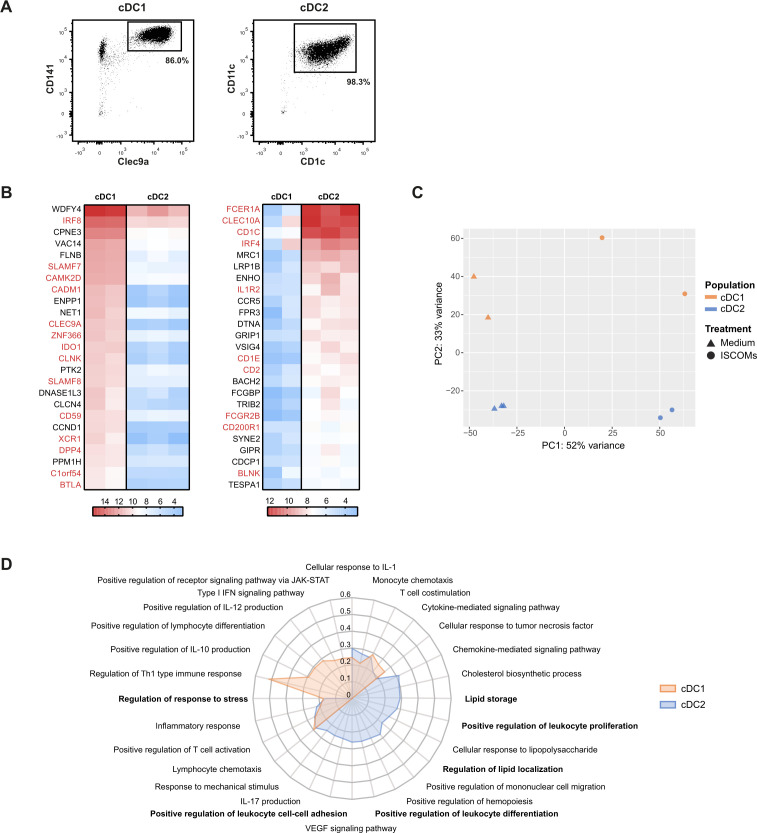
Figure 1.
ISCOMs upregulate distinct pathways in cDC2 and cDC1 subsets. (A) Purity of isolated cDC1s and cDC2s after MACS isolation analyzed by flow cytometry. (B) Top 25 highest expressed genes for cDC1s (left heatmap) and cDC2s (right heatmap) without ISCOMs stimulation based on transformed normalized counts from RNA sequencing. Known genes that are specific for each subset according to literature are depicted in red. (C) Principal component analysis (PCA) for bulk-sequenced cDC1s and cDC2s before and after 8 hours of ISCOMs stimulation. (D) Gene ontology (GO) enrichment for Biological Processes based on DEGs with p<0.05 and fold change ≥2 after stimulation with ISCOMs for 8 hours. Immunologically relevant GO pathways were selected and displayed as a radar plot showing the gene ratio. RNA sequencing for bulk subsets was performed on two to three healthy donors. cDC1, conventional type 1 DC; cDC2, conventional type 2 DC; DEGs, differentially expressed genes; IFN, interferon; IL, interleukin; ISCOMs, immune stimulatory complexes; JAK-STAT, Janus kinases-signal transducer and activator of transcription proteins; MACS, magnetic-activated cell sorting; Th, T helper; VEGF, vascular endothelial growth factor.