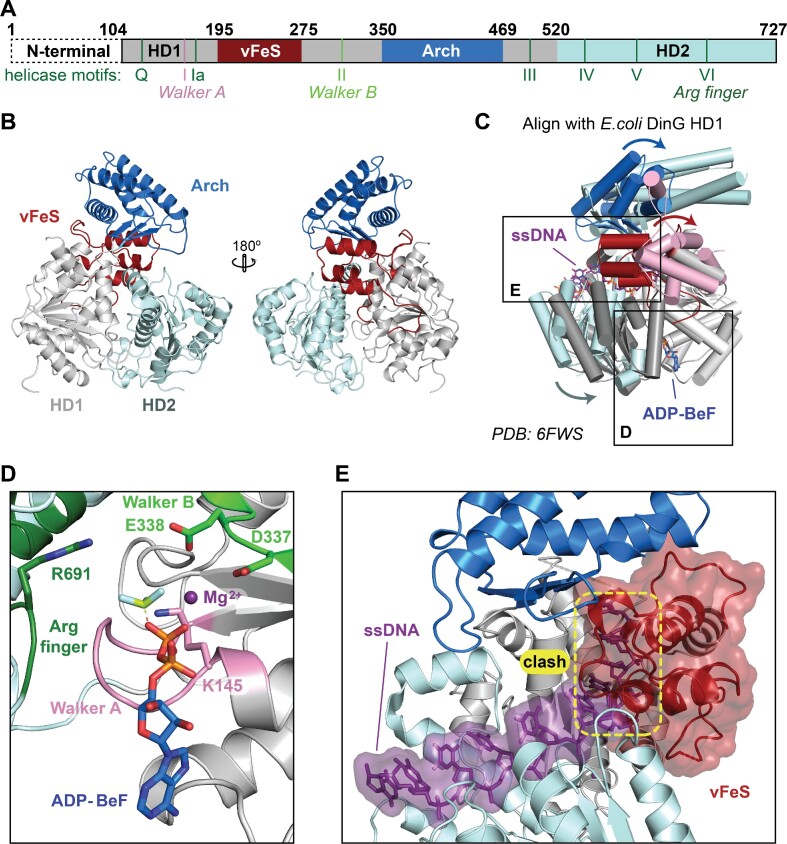
Figure 3.
X-ray crystal structure of CasDinG. (A) Diagram of the CasDinG protein sequence depicting the HD1 (silver) and HD2 domains (pale cyan) with accessory domains vFeS (red) and Arch (blue). The positions of SF2 helicase motifs are indicated under the diagram. The N-terminal domain of CasDinG is indicated but was unresolved in the solved structure. (B) Model of CasDinG highlighting the Arch and vFeS accessory domains. (C) Alignment of the CasDinG model with the E. coli DinG model bound to ssDNA and ADP-BeF3 Mg2+ complex (PDB 6FWS). Positional differences between the CasDinG and E. coli DinG are indicated with arrows. (D) Zoomed in view of the ATP binding pocket with the aligned ADP-BeF3 Mg2+ complex (6FWS) to the CasDinG model. The Walker A (pink) and Walker B (lime) motifs are in a position to coordinate the ADP-BeF3 Mg2+. (E) View of the nucleic acid binding pocket showing the aligned ssDNA (6FWS) within the RecA-like fold of the HD1 and HD2 domains. Highlighted in yellow is a clash between the aligned ssDNA and the CasDinG vFeS domain, indicating the vFeS domain is in a closed position that would have to open to accommodate bound ssDNA.