Figure 4.
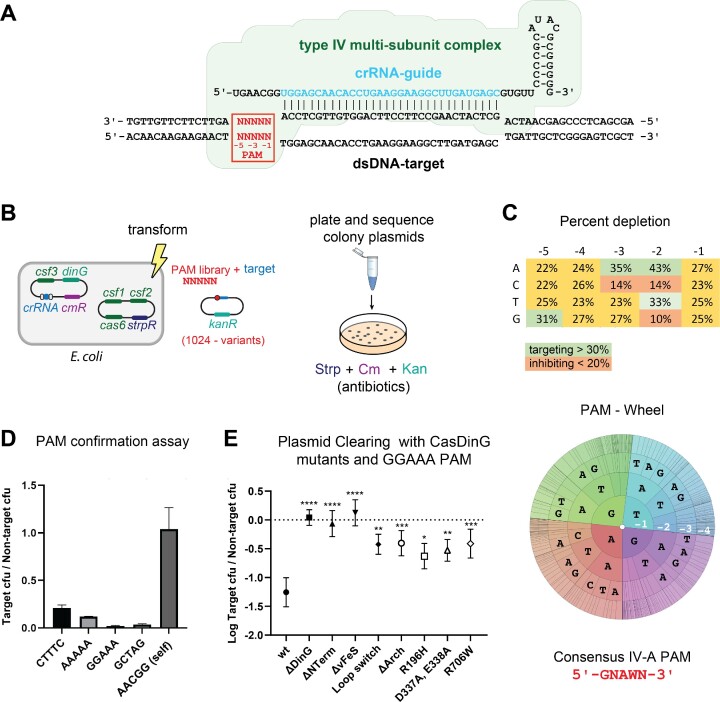
The type IV-A CRISPR system prefers a GNAWN PAM and requires CasDinG accessory domains. (A) Schematic depicting the type IV-A Csf complex binding to a dsDNA target. The location of the PAM is indicated in red. (B) Diagram of how the PAM library was performed. E. coli cells expressing the type IV-A system were transformed with a library of target plasmids adjacent to all possible combinations of PAM sequences. The transformation was grown under antibiotic selection, cells were harvested, and DNA was isolated and deep sequenced. (C) Table (top) and PAM wheel (bottom) showing percent depletion of nucleotides in the PAM assay, highlighting a consensus PAM 5′- GNAWN -3′ sequence. (D) The ratio of target /non-target colony forming units using four different targeting PAM sequences compared to self. (E) Plasmid curing assay with domain and point mutants, demonstrating these domains are essential for type IV-A CRISPR immunity.