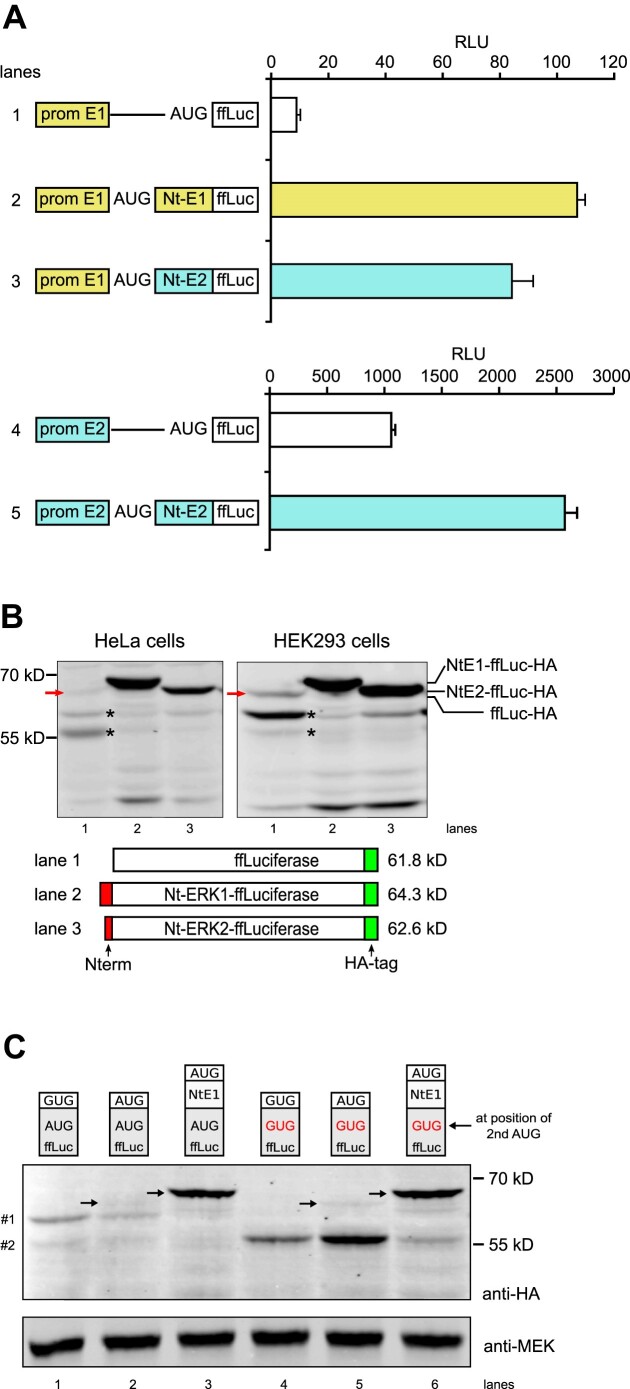
Figure 3.
Firefly luciferase (ffLuc) as a reporter to study ERK Nt action on start codon selection. (A) Normalized ffLuc activity (RLU) from extracts of NIH3T3 cells transfected with plasmids containing the erk1 promoter and the 5′-UTR (prom E1), in the absence (lane 1) or presence of ERK1 or ERK2 Nt moieties fused to ffLuc (lanes 2 and 3). Transfection of plasmids with the erk2 promoter and 5′-UTR (prom E2) in the absence (lane 4) or presence of ERK2 Nt fused to ffLuc (lane 5) (n = 3, representative of >3 experiments). (B) Western blot analysis of extracts from HeLa cells and HEK293 cells transfected with various constructs, all in the context of the erk1 promoter and 5′-UTR. The presence of Nt domains (red), the position of the HA tag (green) and the predicted molecular weight of full-length proteins are indicated. Detection of HA-tagged luciferase was performed using an anti-HA antibody. The red arrow corresponds to ffLuc-HA at the expected size (lane 1); asterisks (*) indicate leaky scanning bands. (C) Western blot analysis from HEK293 cells transfected and processed as in (B). The first three lanes express WT-ffLuc-HA while the last three lanes express ffLuc-HA with its second in-frame AUG mutated to GUG. In lanes 1 and 4, ffLuc lacks the first AUG while in lanes 3 and 6 the ERK1 Nt domain was fused to ffLuc. Small molecular weight polypeptides expressing the HA-tag are indicated on the left (#1 and #2). Arrows indicate functional ffLuc-HA at the expected size.