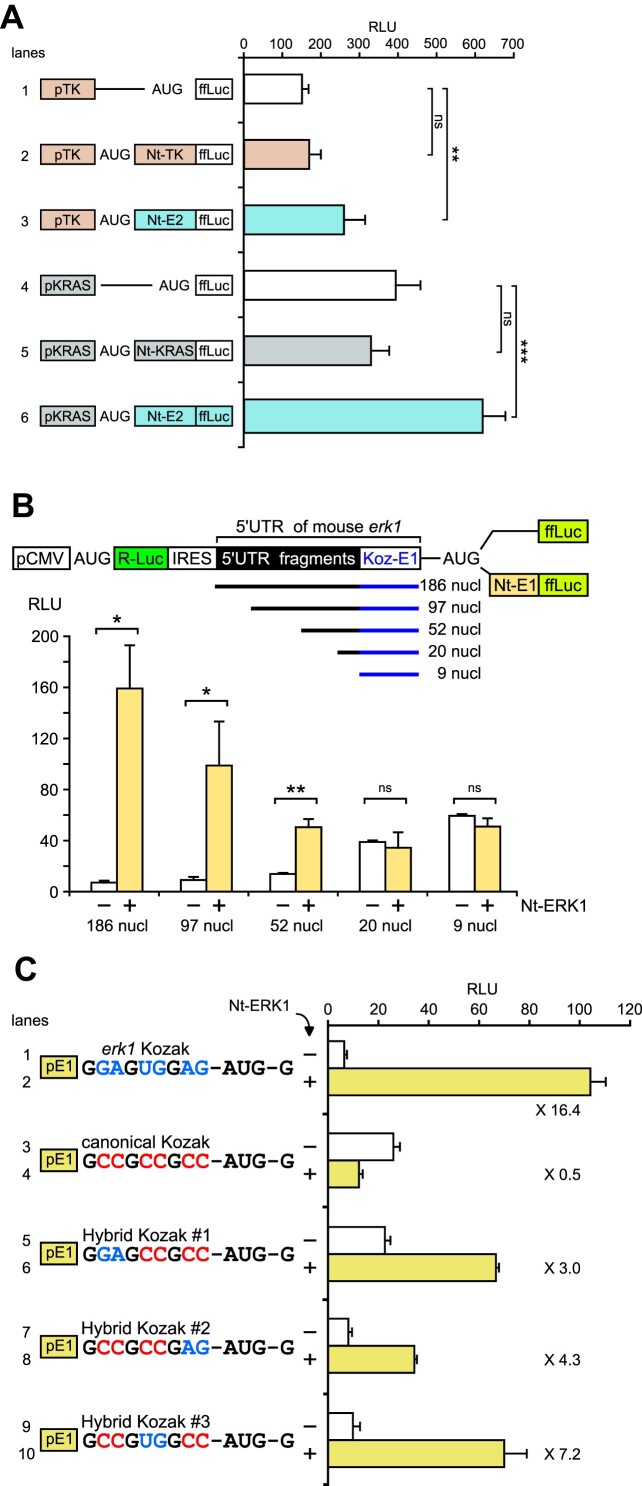
Figure 5.
Analysis of 5′-UTR features that cooperate with the ERK Nt to determine start codon selection. (A) RLU from extracts of NIH3T3 cells transfected with plasmids bearing HSV-TK (orange) or KRAS (gray) promoters and the 5′-UTR. ffLuc was expressed directly behind promoters (white bars) or fused to their homologous Nt domain (orange and gray) or ERK2 Nt moiety (blue) (n = 6, ns = not significant, **P < 0.01 or ***P < 0.001 bilateral Welch's t-test, representative of three experiments). (B) NIH3T3 cells transfected with bicistronic constructs depicted above the graph. The CMV promoter expresses one mRNA, with R-Luc normalizing ffLuc (RLU). The second cistron, ffLuc, is fused (yellow bars) or not (white bars) to the ERK1 Nt. Upstream of the erk1 Kozak AUG sequence, fragments of the mouse erk1 5′-UTR are cloned with the indicated sizes (0–177 nucleotides, deletions from the 5′ side) (n = 3, ns = not significant, *P < 0.05 or **P < 0.01 bilateral Welch's t-test, representative of three experiments). (C) RLU from NIH3T3 cells transfected with plasmids harboring distinct Kozak sequences in the context of promoter erk1 and its 5′-UTR (pE1). ffLuc is fused (yellow bars) or not (white bars) to the ERK1 Nt, and fold induction is indicated on the graph near the yellow bars. Lines 1 and 2 contain erk1 Kozak and lines 3 and 4 the canonical Kozak sequence. From lines 5–10, nucleotides of Kozaks were swapped as indicated. Guanine nucleotides shared by erk1 and the canonical Kozak context are in black, nucleotides specific to the erk1 Kozak context in blue and those specific to the canonical Kozak context in red (n = 3, representative of three experiments).