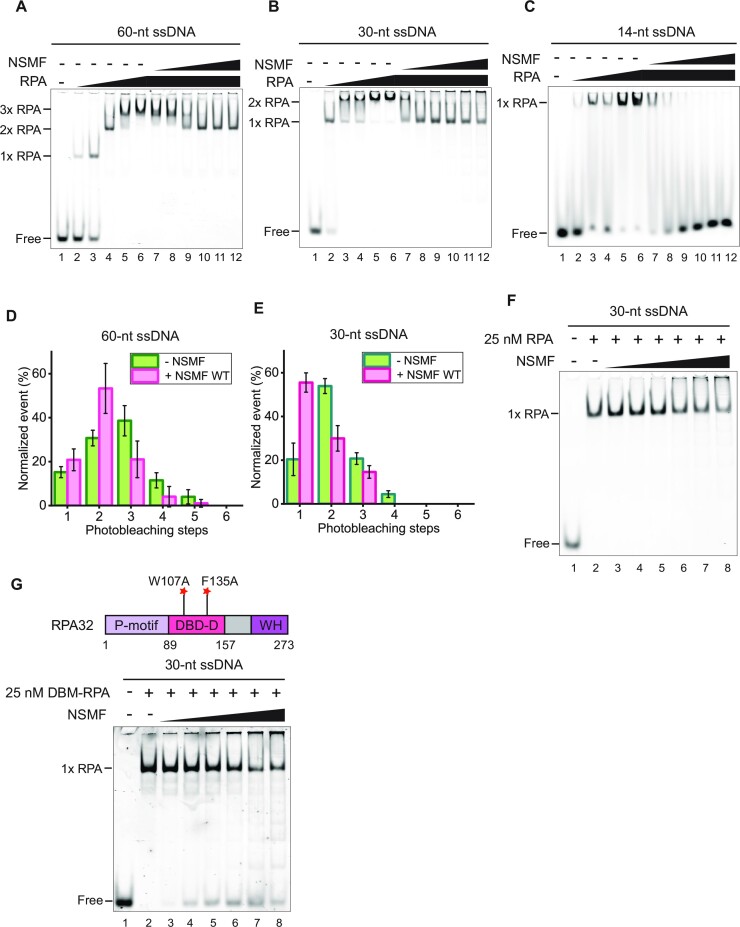
Figure 3.
NSMF modulates the RPA binding mode. (A–C) EMSA for NSMF with RPA bound to (A) 60-nt, (B) 30-nt, and (C) 14-nt ssDNA. RPA (0, 25, 75, 100, 125, and 150 nM) was titrated with 10 nM of each ssDNA. RPA (150 nM) was titrated with NSMF (0, 10, 20, 40, 80, 140, and 200 nM). (D, E) Histograms for photobleaching events of RPA-eGFP bound to (D) 60-nt and (E) 30-nt ssDNA in the presence (magenta) or absence (green) of NSMF. The addition of NSMF reduced the number of bound RPA molecules. More than 100 molecules were analyzed for each dataset. Error bars represent the standard deviation in triplicate. (F) NSMF (0, 10, 20, 40, 80, 140, and 200 nM) was titrated with 25 nM RPA bound to 30-nt ssDNA. At 25 nM, RPA had a single binding mode (30-nt mode) without a band shift. (G) Top: the domain structure for the DNA binding defective mutant of RPA (DBM-RPA), in which Trp107 and Phe135 were both replaced by Ala. Bottom: EMSA for NSMF with DBM-RPA and 30-nt ssDNA. DBM-RPA (25 nM) was bound to 10 nM 30-nt ssDNA and then titrated with NSMF (0, 10, 20, 40, 80, 140, and 200 nM).