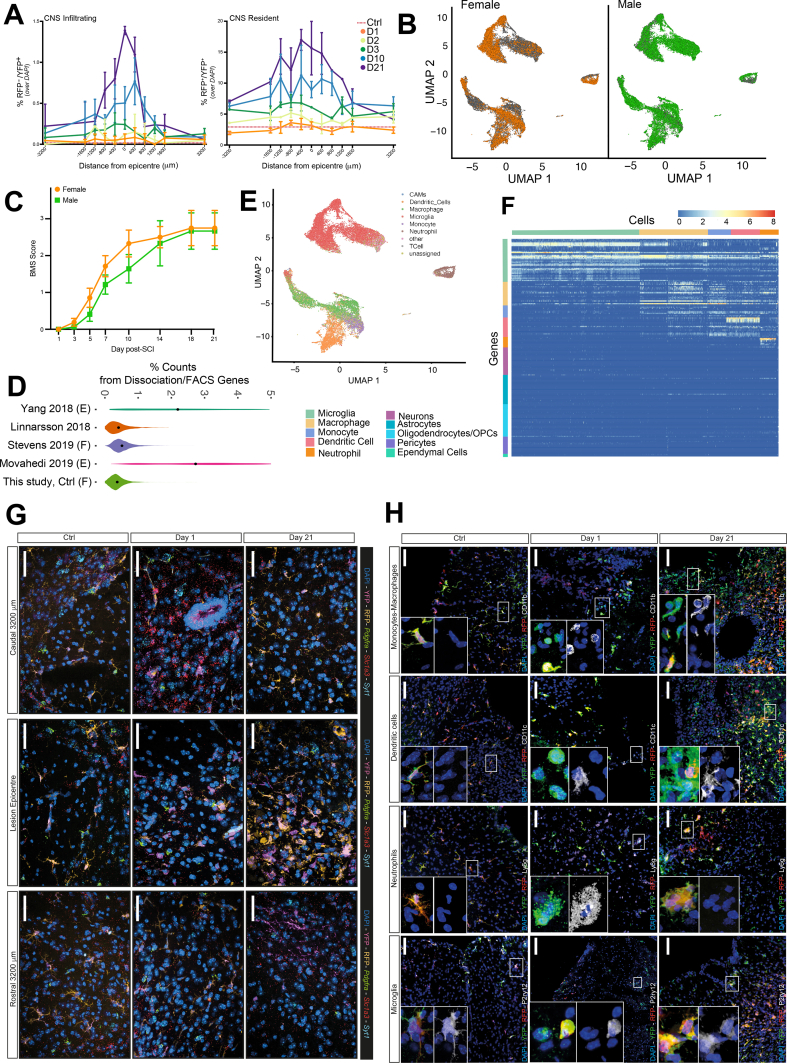
S. Fig. 2.
Quality controls. A, Quantification of RFP-/YFP+ infiltrating myeloid cells (left) or RFP+/YFP+ CNS resident myeloid cells (right) as a proportion of DAPI+ at set distances rostral (+) or caudal (-) of the lesion epicentre. Data are mean % (± SD) and have been collected from n= 3 mice per time point. B, UMAPs of cells from male vs. female mice. C, Basso Mouse Score (BMS) from male and female mice. Data are mean numbers (± SEM) and have been collected from n ≥ 4 mice per time point. D, Violin plots showing the percentage of all UMI counts originating from dissociation and FACS associated genes, per Ctrl MG cell, across several droplet-based scRNAseq studies after applying the same quality control metrics. Black dots are the mean. F= FACS, E= enzymatic (vs. mechanical) dissociation. E, UMAP of the entire dataset, coloured by CellAssign annotation, before manual assignments. F, Heatmap of the log2-transformed counts of canonical marker genes for CNS-associated cell types. G, smFISH analysis of the expression of cell-type markers Pdgfra (oligodendrocyte progenitor cells), Slc1a3 (astrocytes), and Syt1 (neurons) from Ctrl, 1 dpi, and 21 dpi. H, Confocal microscopy images of the expression of P2ry12, Ly6g, CD11c and CD11b by MG, NPss, DCs and monocytes-macrophages, respectively, at the lesion epicentre. Nuclei were stained with DAPI. Scale bars: 60 μm.