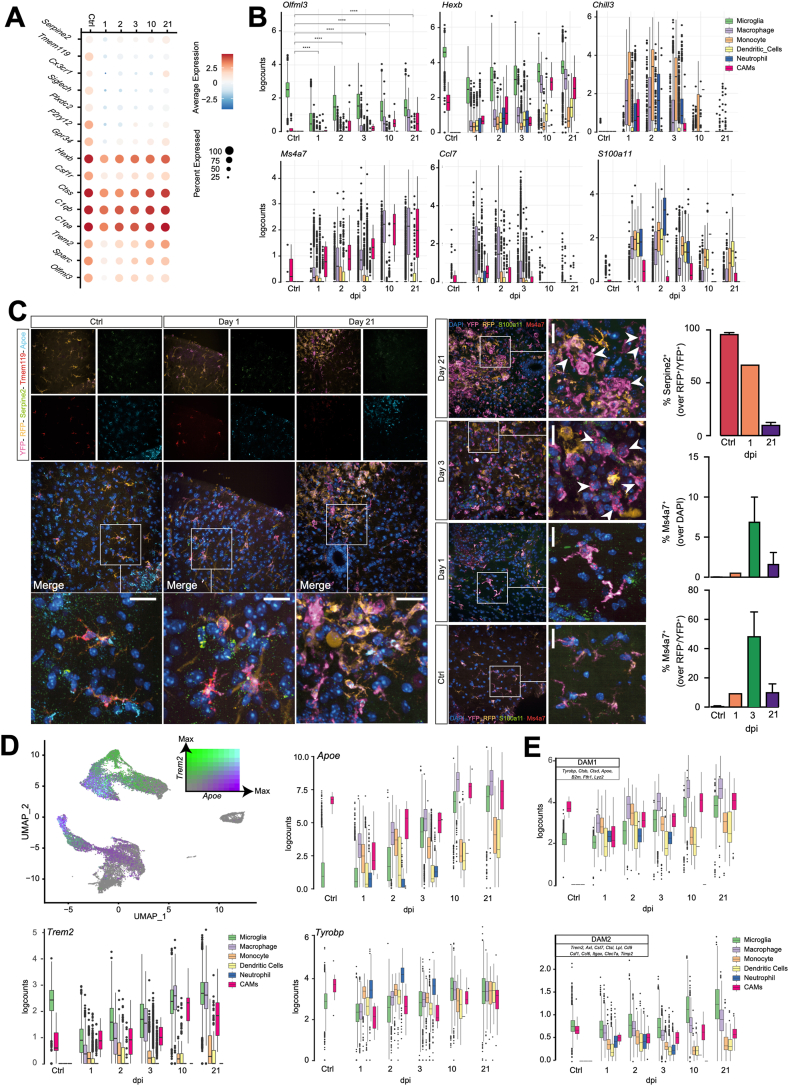
Fig. 2.
Validation against published myeloid cell studies via both scRNAseq and smFISH. A, Seurat dot plot showing canonical MG marker expression by dpi. Scale bar is the natural log of the average log2-transformed counts. B, Boxplots by cell type and time point of the log2-transformed expression of selected genes. For Olfml3: Welch t-test with Benjamini-Hochberg post-hoc correction.ns p ≥ 0.05; *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001). Boxplots for every gene in this dataset can be found on the SCI Myeloid Cell Atlas shiny app (https://marionilab.cruk.cam.ac.uk/SCI_Myeloid_Cell_Atlas/). C, smFISH data depicting the expression of Tmem119, Serpine2 and Apoe from Ctrl, 1 dpi, and 21 dpi (left) and Ms4a7 and S100a11 from Ctrl, 1, 3, and 21 dpi (right). The bar graph shows the quantification of Ms4a7+ and Serpine2+ cells. Arrowheads indicate Ms4a7+/RFP−/YFP+ cells. Data are mean % (±SD) from n = 2 mice per time point. Nuclei were stained with DAPI. Scale bars: 20 μm. D, UMAP coloured by the scaled log2-transformed counts of Apoe and Trem2. Boxplots by time and cell type demonstrating the upregulation of TREM2-induced APOE pathway-associated genes Apoe, Trem2, and Tyrobp upon SCI. E, Boxplots by time point coloured by cell type, quantifying the average log2-transformed counts of DAM1-vs. DAM2-associated genes expressed per cell.