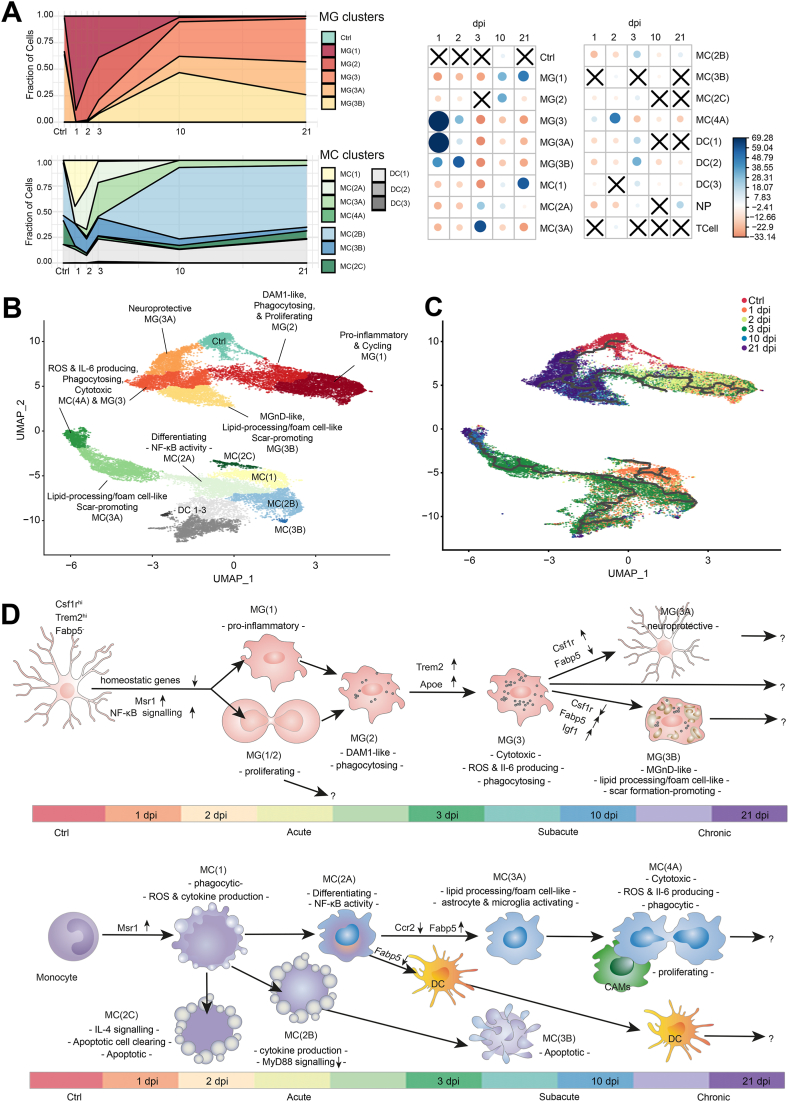
Fig. 3.
Dynamic trajectories of myeloid cells post-SCI. A, Breakdown of the MG and MC clusters by the fraction of cells from each time point (left), and correlation plot of the Pearson's chi-squared residuals (right). The size of each circle is proportional to the absolute value of the standardized chi-square residual. Blue signifies a positive standardized chi-square residual, while red signifies the opposite. “X” represents a non-significant contribution. B, UMAP coloured by cluster, as determined by Leiden community detection using Monocle 3. Clusters are named based on their majority cell-type and their position in the trajectory. The UMAP is superimposed with the unsupervised reversed graph embedding-derived trajectory inferred via Monocle 3. C, UMAP coloured by time post-SCI, giving directionality to the trajectory. Trajectories of B are represented by the black lines. D, Proposed map of MG (top) or MC (bottom) states over time.