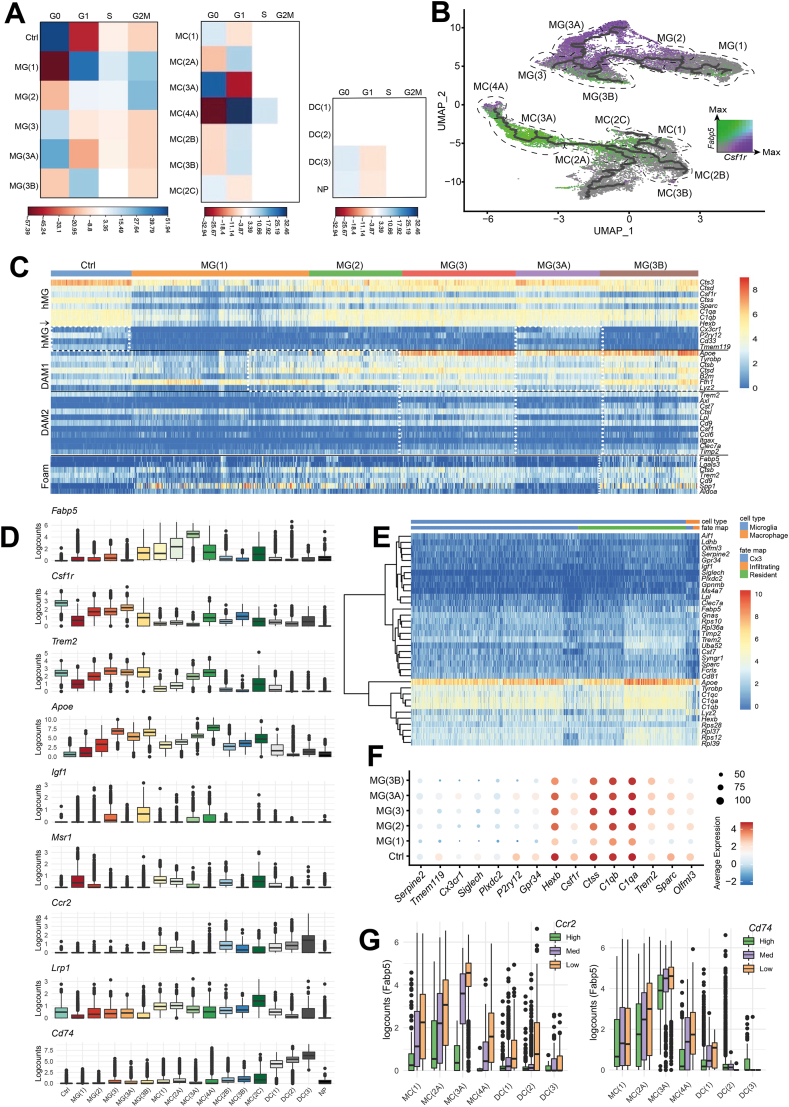
Fig. 4.
Clusters represent the cell states of myeloid cell subpopulations post-SCI. A, Correlation plot of the Pearson's chi-squared residuals between cell cycle phase (columns) and MG/MC cluster (rows) membership: MG Pearson's Chi-squared test X-squared = 7417.2, df = 18, p-value <2.2e-16 or MC Pearson's Chi-squared test X-squared = 2414, df = 33, p-value <2.2e-16. B, UMAP coloured by the scaled log2-transformed counts of Fabp5 (green) or Csf1r (purple). Dotted lines depict relevant clusters. C, Heatmap of homeostatic, DAM-associated, and foam cell genes in MG across the different clusters. Profiles are separated by solid black line. Each row is a gene, and each column is a cell. Dotted white boxes highlight MG that fit the designated profile. hMG = homeostatic MG, hMG↓ = homeostatic MG genes that are downregulated in the DAM1 signature. D, Boxplots of the log2-transformed counts of selected genes by cluster. E, Heatmap log2-transformed counts for the top 20 differentially expressed genes for MG(A2) plus Serpine2 and Gpnmb. Cells (columns) are labelled by their fate mapping status. F, Seurat dot plot showing canonical MG marker expression by cluster. Scale bar is the natural log of the average log2-transformed counts. G, Boxplot of the Fabp5 expression across the MC(1), MC(A), and DC clusters. Left: Cells are binned by Ccr2 expression levels; High ≥ 10 counts; 1 < Med <10; Low. Right: Cells are binned by Cd74 expression levels; High ≥ 20 counts; 1 < Med <20; Low =< 1.