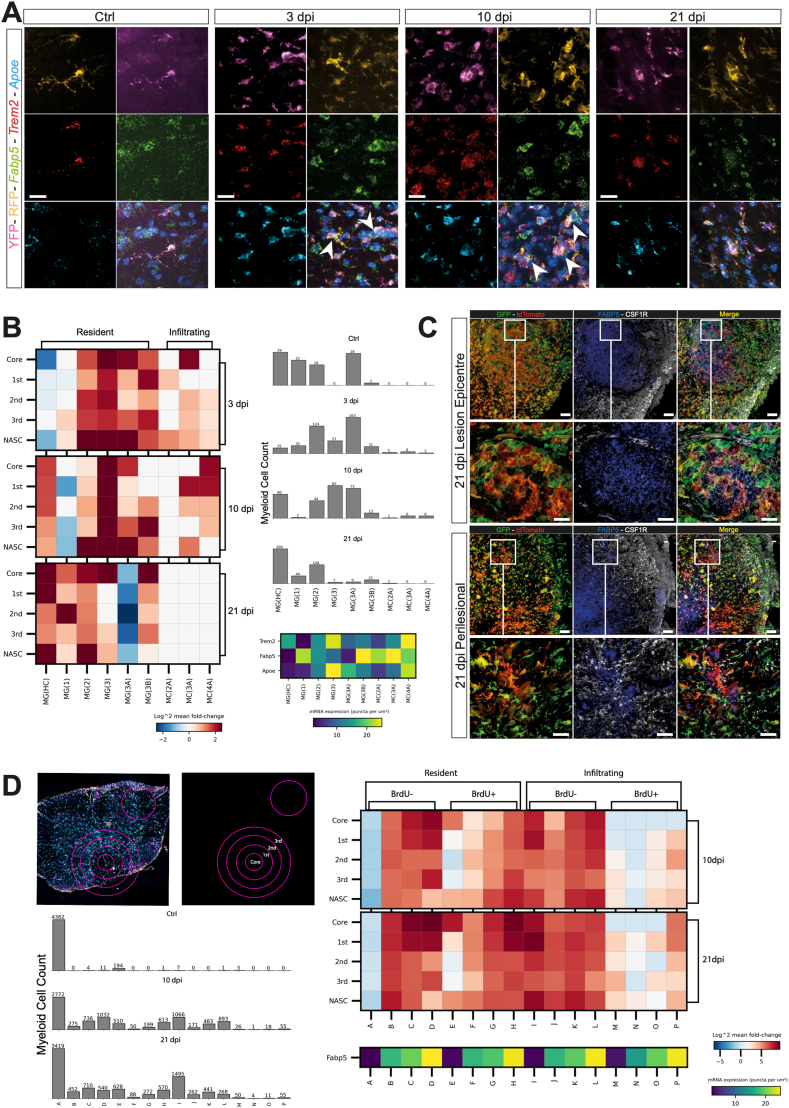
Fig. 5.
smFISH and confocal immunofluorescence microscopy validation and spatial context. A, Expression of Fabp5, Trem2 and Apoe. Nuclei were stained with DAPI. Arrowheads indicate Fabp5+/RFP+/YFP+ cells. Scale bars: 20 μm. B, Heatmap of smFISH data depicting the spatial distribution of the scRNAseq clusters at the lesion epicentre and in the perilesional cord, as in the representative images. Data are presented as mean fold change over Ctrl. Quantification of cells per bin is shown in the bar graphs on the right. Bottom right is the expression level of the three smFISH probes, Apoe, Fabp5, and Trem2 defining each cluster. C, Confocal microscopy images of the expression of FABP5 and CSF1R at the lesion epicentre (top) and in the perilesional cord (bottom) at 21 dpi. Nuclei were stained with DAPI. Scale bars: main panels: 60 μm; insert panels: 20 μm. D, Heatmap of smFISH data from the BrdU, acute proliferation experiment at the lesion epicentre and in the perilesional cord, as in B. Data are presented as mean fold change over Ctrl. Bottom left heatmap of smFISH data depicts the spatial distribution of binned cells and the expression level of Fabp5 defining each bin. Data are binned by BrdU expression (±) and Fabp5 expression (-/lo/med/hi). Quantification of cells per bin is shown in the bar graphs on the right. Data are mean % (±SD) from n = 2 mice per time point.