Fig. 2. Overview of computational workflow for anchor position prediction.
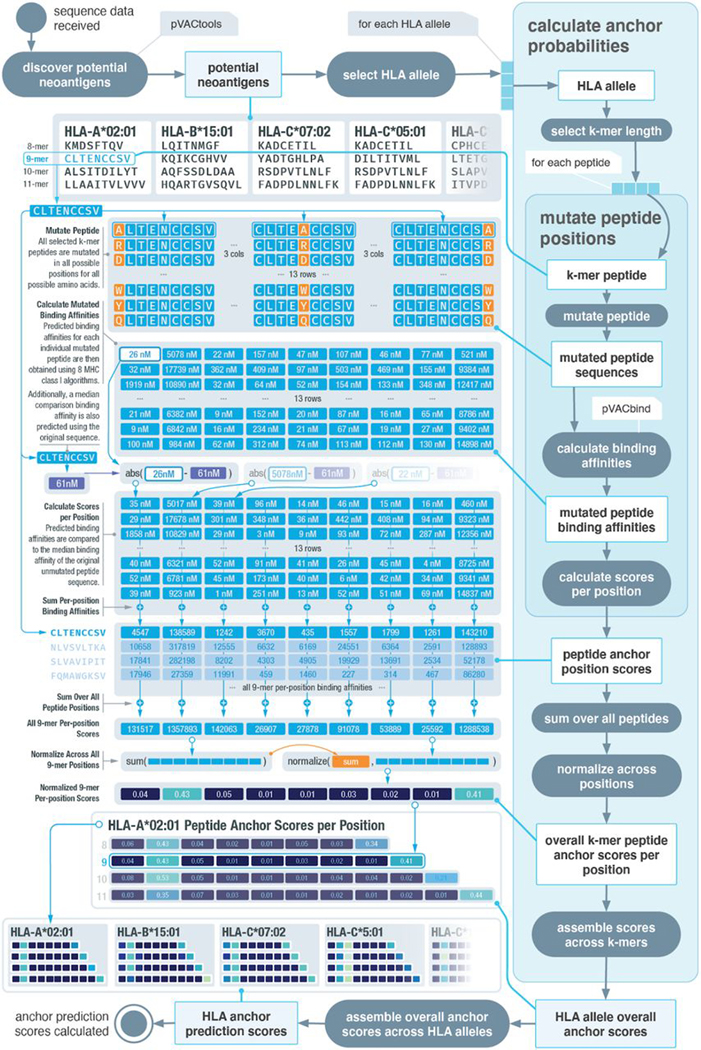
Schematic of our computational workflow to simulate the effect of mutation position on MHC binding affinities of peptides for individual HLA alleles. HLA and peptide pairings are selected from a reference dataset of putative strong binders. All possible amino acid changes are applied to all possible positions, and the impact on binding affinity is assessed. An overall peptide anchor score is calculated for each position for all HLA-peptide length combinations. Higher scores indicate greater likelihood that a particular position in the peptide acts as an anchor residue for a given HLA.
