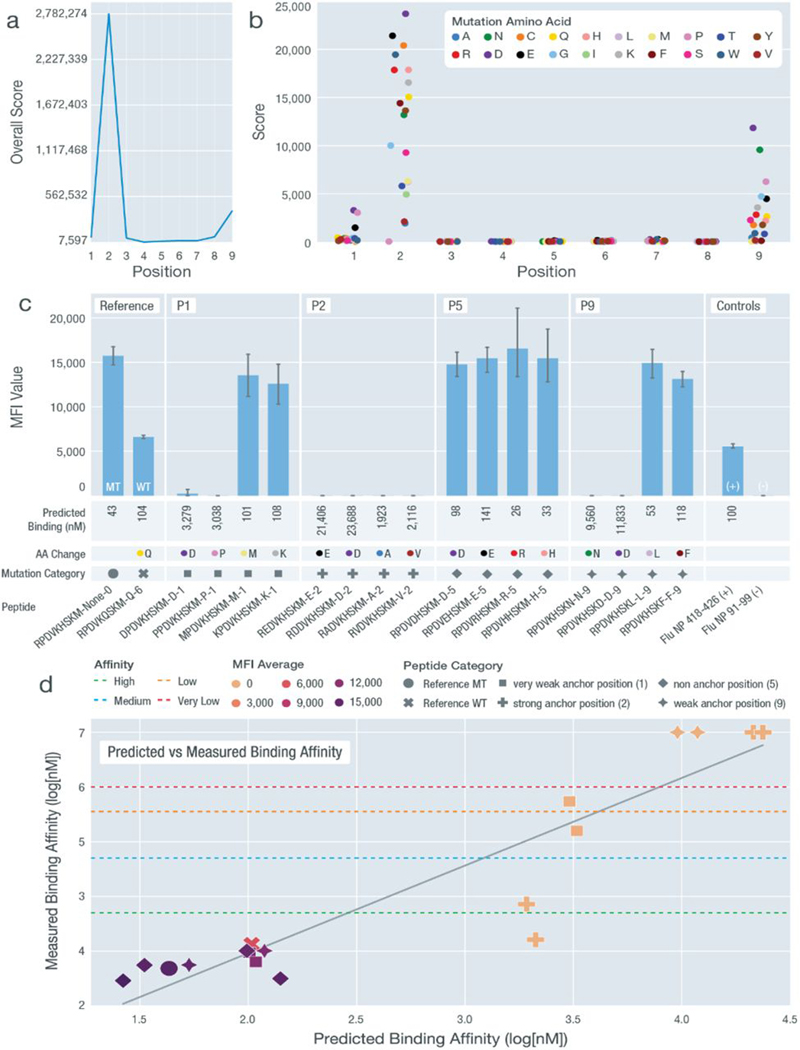
Fig. 5. Experimental validation of anchor pattern for HLA-B*07:02.
Validation results for HLA-B*07:02 with a predicted 2S-9W anchor pattern using peptide sequence RPDVKHSKM. (A) Overall anchor prediction scores for each position of 9-mer peptides when binding to HLA-B*07:02. Higher scores indicate a higher probability of acting as an anchor. (B) Binding affinity changes (y axis) plotted for each position of the specific 9-mer peptide RPDVKHSKM. Each peptide position was mutated to 19 other amino acids to evaluate influence on binding affinity. (C) MFI values measured from cell stabilization assays are plotted for both unmutated and mutated peptides. Peptides are marked by their mutation positions (P1, P2, P5, and P9), predicted binding affinity values, amino acid changes [color coordinated with (B)], and mutation category [shape coordinated with (D)]. (D) Predicted binding affinity scores (log10[nM]) plotted against measured binding affinity values (log10[nM]) from IC50 binding assays. Binding categories based on measured binding affinity values are marked using horizontal lines. MFI values are overlaid using heatmap coloring for all data points. Peptide categories are marked using different shapes [coordinated (C)]. All nonbinders were plotted with a measured binding affinity value of 7.