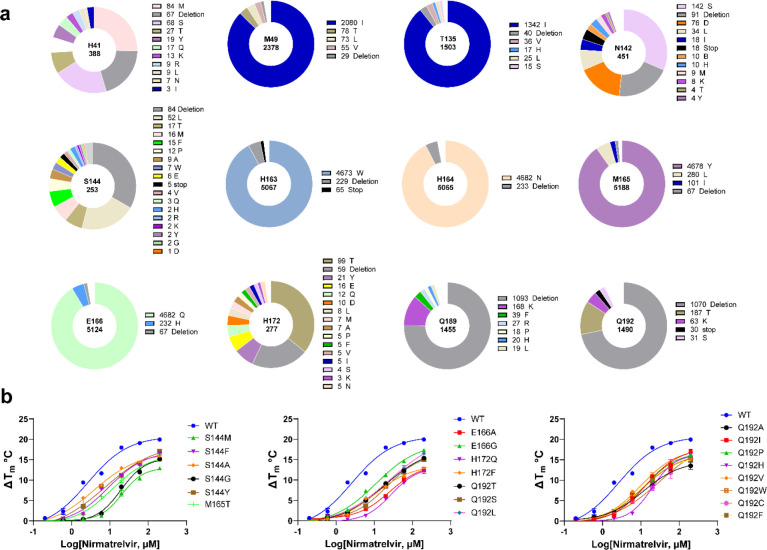
Figure 2.
SARS-CoV-2 Mpro mutants characterized in this study. (a) Mutations at 12 residues located at the nirmatrelvir-binding site. Sequence data were obtained from CoVsurver of the GISAID, developed by A*STAR Bioinformatics Institute (BII), Singapore. A total of 5 420 461 mutations of Nsp5 (Mpro) were obtained from the database as of July 7, 2022. Occurrence of total mutations for each amino acid residue is labeled in the center of each pie chart. The occurrences of specific mutations are labeled on the right of its affiliate pie chart. (b) Characterization of nirmatrelvir resistance against Mpro mutants by the thermal shift assay. The results are the average of duplicates.