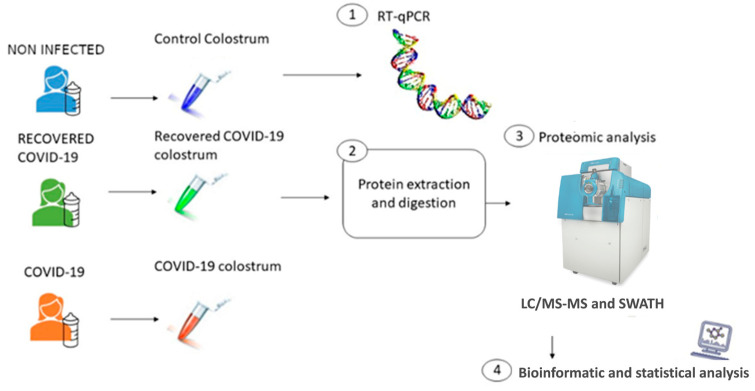
Figure 1.
Schematic representation of the study design. Colostrum samples were collected from active COVID-19 women (n = 3), recovered (n = 4) and noninfected women (n = 5). The workflow for processing the omics study was as follows: Firstly, RT-qPCR was performed to determine the presence or absence of viral RNA. Secondly, protein extraction and digestion for SWATH-MS and proteomic analysis was performed. Finally, proteomic results were analyzed using statistical and bioinformatics analysis.