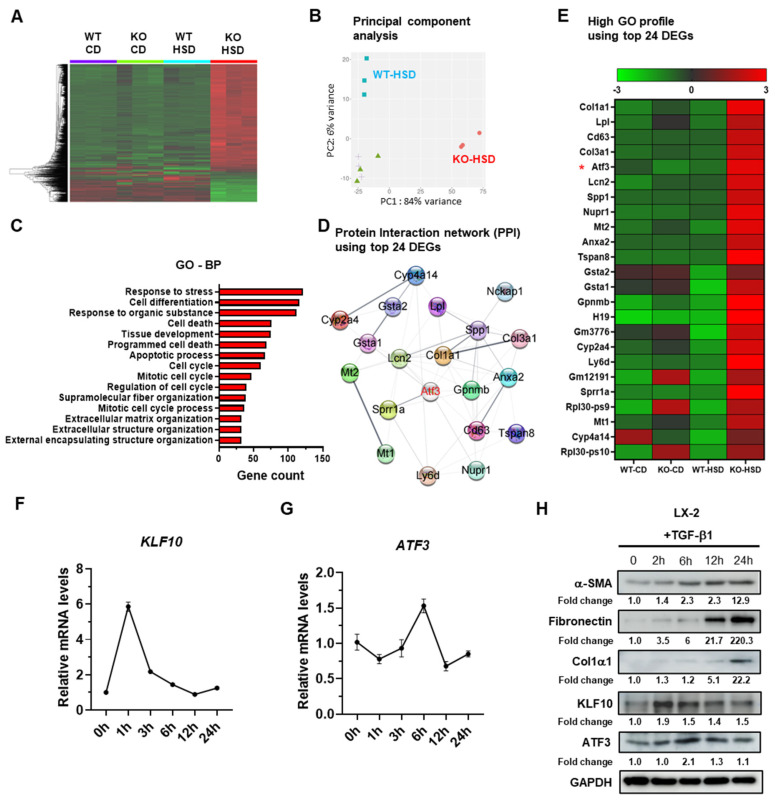
Figure 1.
ATF3 is upregulated in the fibrotic liver of HSD-fed KLF10 KO mice and in activated HSCs. (A) Different genes in the livers of WT and KLF10 KO mice fed a CD or HSD. (B) Principal component (PC) analysis. (C) The 15 most significant GO-BP pathways of the differentially expressed genes between HSD-fed WT and KLF10 KO mice. GO-BP, Gene ontology-biological process. (D) Protein–protein interactions (PPI) generated by Cytoscape. (E) Heatmap of the top 24 upregulated genes between HSD-fed KLF10 KO and WT mice. ATF3 in the pathway of high GO profile and PPI analysis is marked in red. *, ATF3 as a potential target gene of KLF10 during liver fibrosis. The relative mRNA levels of KLF10 (F) and ATF3 (G) were determined by qPCR. LX-2 cells were treated with TGF-β1 for the indicated times. The target gene expression was normalized to the expression of cyclophilin and expressed as means ± SEM. (H) Western blot applied to LX-2 cells treated with TGF-β1 (10 ng/mL) for the indicated times. The levels of α-SMA, fibronectin, Col1α1, KLF10, and ATF3 were determined. GAPDH was used as an internal control. The protein band of Western blotting was quantified by ImageJ and normalized using GAPDH.