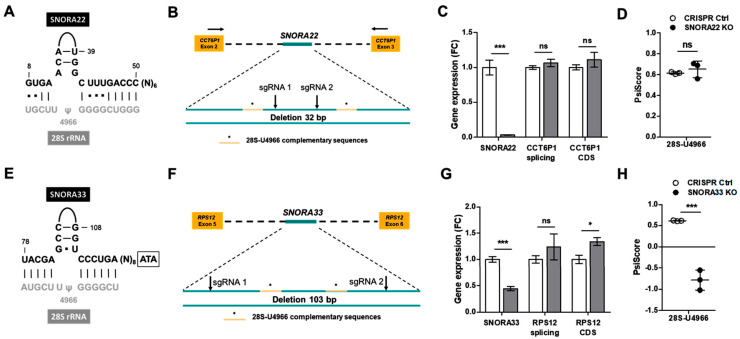
Figure 2.
CRISPR/Cas9-mediated SNORA33 depletion leads to the loss of pseudouridylation at 28S-U4966. (A,E) Two box H/ACA snoRNAs, SNORA22 and SNORA33, are predicted to guide the ψ of 28S-U4966. (B,F) A scheme of the double sgRNA CRISPR/Cas9-mediated approach to generate SNORA22 and SNORA33 SW1353 KO cell pools. Top: Exons of host genes CCT6P1 and RPS12 flanking the intron-encoded SNORA22 and SNORA33, respectively. Bottom: Scheme of SNORA22 and SNORA33 sequences with indicated sgRNA1 and sgRNA2 cleavage sites and rRNA complementary sequences. The cell pool of GFP targeting CRISPR/Cas9-treated cells was used as a control (CRISPR Ctrl). (C) Expression levels of SNORA22, expression and splicing (exons 2 and 3) of its host gene CCT6P1 measured by RT-qPCR (n = 3). (G) Expression levels of SNORA33, expression and splicing (exons 5 and 6) of its host gene RPS12 measured by RT-qPCR (n = 3). Expression levels were normalized to the reference gene (PPIA) expression and plotted as fold change to CRISPR Ctrl. Data were analyzed by unpaired t-test with the assumption of a normal distribution. (D,H) Pseudouridylation levels of 28S-U4966 (n = 3) measured by HydraPsiSeq and analyzed by unpaired t-test with the assumption of a normal distribution of data. Negative PsiScore for 28S-ψ4966 in the case of SNORA33 KO cell pool indicates even more intensive hydrazine cleavage at this position compared to other neighboring U residues. Ns – not significant, * p < 0.05, *** p < 0.001.