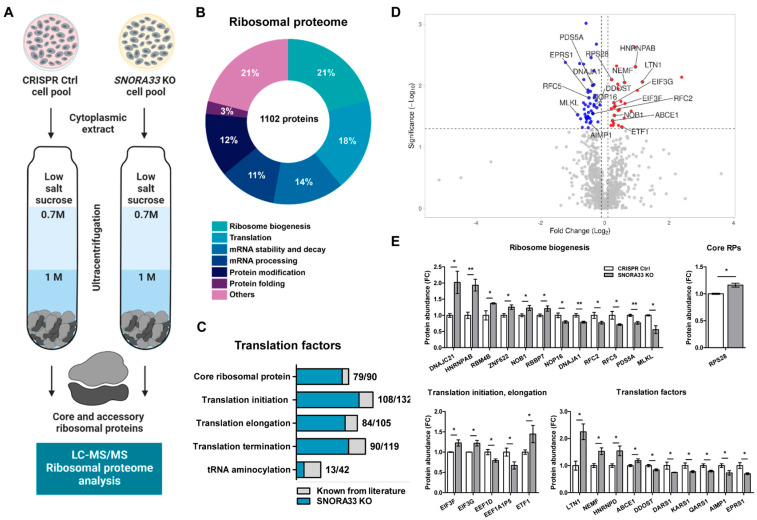
Figure 5.
Depletion of SNORA33 affects ribosomal protein composition. (A) The scheme of the experimental set-up. Cytoplasmic extracts of SNORA33-depleted and CRIRP Ctrl cells (n = 3) were depleted of mitochondria and nuclei and ultracentrifuged through the sucrose cushions. Ribosomal pellets were resuspended and analyzed by label-free LC-MS/MS. (B) A pie chart of all identified ribosomal proteins and their functional allocation. (C) Translation factors identified in the ribosomal proteome. (D) A volcano plot of all identified ribosomal proteins. Normal distribution of the data and equal variance of populations was assumed. Statistical significance was assessed by one-way ANOVA. The dotted lines represent cut-off values (FC ≥ 1.1; p < 0.05). Ribosomal proteins significantly less or more abundant in ribosomes of SNORA33-depleted cells are in red and blue, respectively. (E) Protein abundance of selection of differentially abundant core and accessory ribosomal proteins in SNORA33 KO cell pool. Data are presented as fold changes to CRISPR control. * p < 0.05, ** p < 0.01.