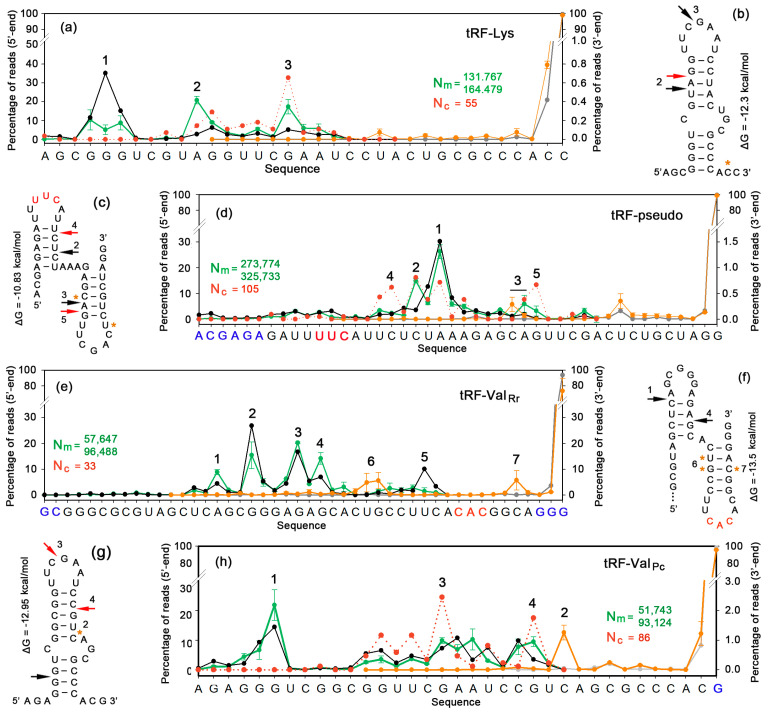
Figure 6.
Synthetic analogs of model tRFs are cleaved by nucleases in the culture medium of growing E. coli, and their fragments penetrate into the cells. (a,d,e,h) The distribution of the 5′- and 3′-ends of synthetic analogs (black and gray plots, respectively) and culture media-derived sequences (green and orange plots, respectively). The dotted plots show the distribution of 5′-ends for reads detected within E. coli cells. Nm is the number of tRFs found in milieu from 2 experiments. Nc is the number of such reads found in pooled datasets received in the same experiments inside bacterial cells. (b,c,f,g) Structural models of tRFs. Arrows indicate preferred nuclease cleavage sites generating 5′-ends in culture medium (black) and within recipient cells (red). Asterisks show 3′-terminal nucleotides of reads predominantly accumulated in the milieu.