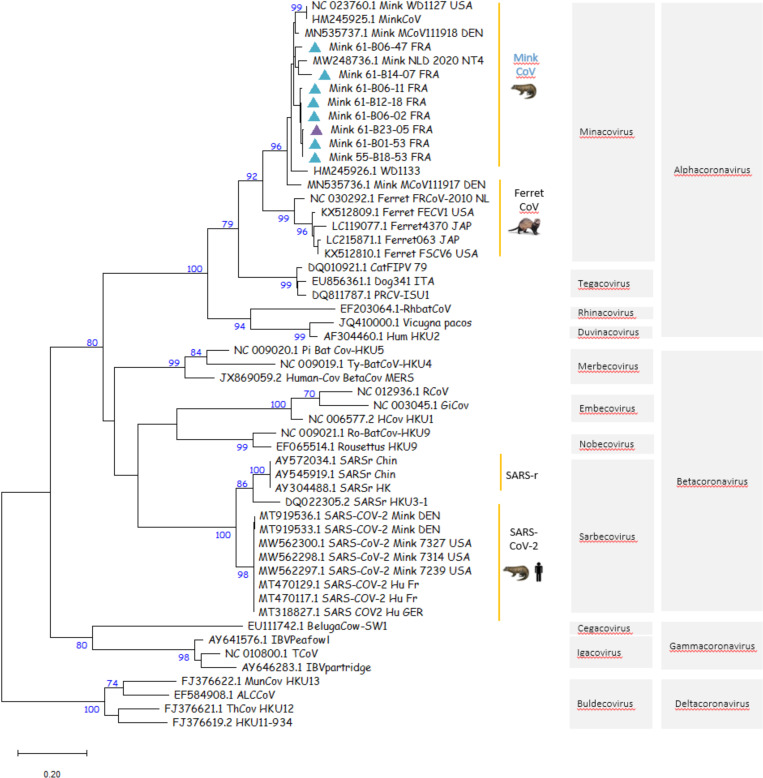
Fig 3. Maximum likelihood (ML) phylogeny inferred with eight mink coronavirus consensus sequences from France and 45 representative GenBank sequences including alphacoronaviruses (n = 25), betacoronaviruses (n = 20) with mink and human SARS-CoV-2 sequences, gammacoronaviruses (n = 4) and deltacoronaviruses (n = 4).
Bootstrap values above 70% were considered as statistically significant. Pharyngo-tracheal swabs and feces infected by the mink coronavirus are represented on the tree by a turquoise or purple triangle, respectively. The eight partial pol gene consensus sequences included in the ML tree are accessible in GenBank under accession numbers: Mink_61-B14-07_FRA: ON985270; Mink_61-B12-18_FRA: ON985271; Mink_61-B06-47_FRA: ON985272; Mink_61-B06-11_FRA: ON985273; Mink_61-B06-02_FRA: ON985274; Mink_61-B01-53_FRA: ON985275; Mink_55-B18-53_FRA: ON985276; Mink_61-B23-05_FRA: ON985277.