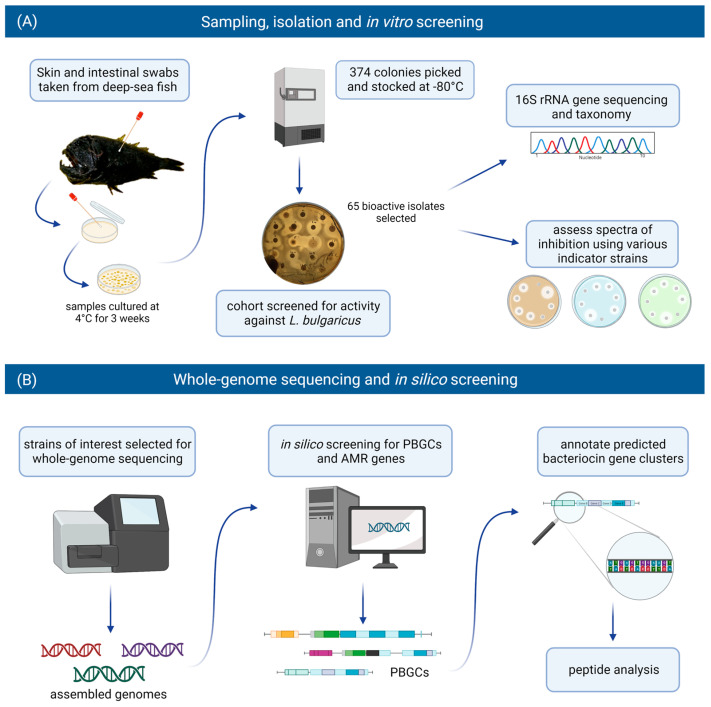
Figure 1.
The in vitro and in silico-based approaches used in this study to assess the bacteriocinogenic potential in the deep-sea fish microbiome. (A) Swabs were taken from the skin and intestines of deep-sea fish. Samples were cultured at 4 °C for three weeks, and 374 isolates were recovered. A cohort of these isolates were screened for activity against Lactobacillus delbrueckii subsp. bulgaricus. Sixty-five bioactive isolates were identified and selected for further in vitro screening and identification through 16S rRNA gene analysis. (B) Strains of interest were selected for whole-genome sequencing and subsequent in silico screening for bacteriocin/RiPP genes, putative biosynthetic gene clusters (PBGCs) and antimicrobial resistance (AMR) genes. Predicted bacteriocin gene clusters were annotated, and the bacteriocin peptide precursors were analysed (figure created with BioRender.com).