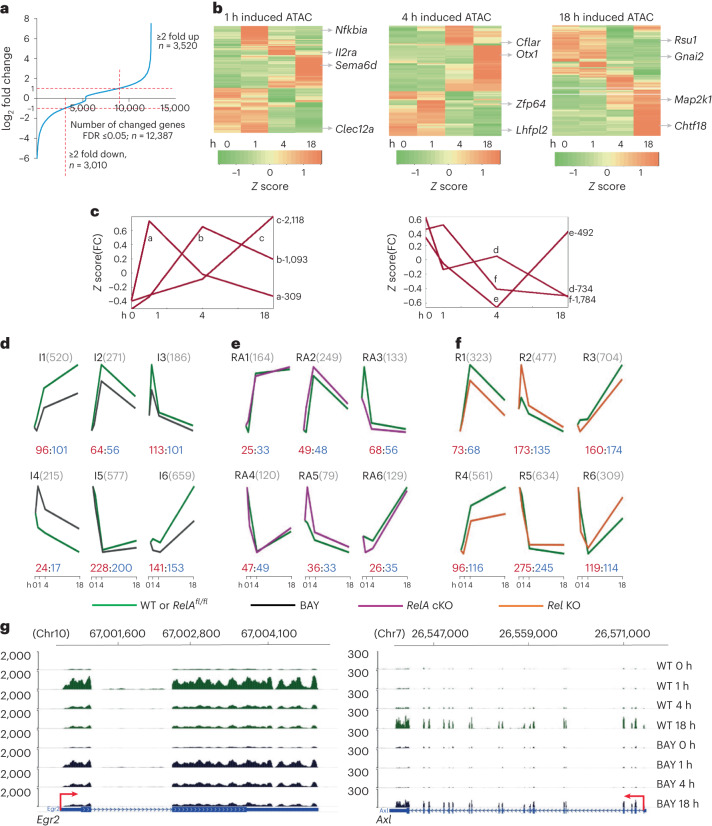
Fig. 3. NF-κB-dependent transcriptional responses in activated B cells.
a–g, Differential gene expression was analyzed using EBSeq (FDR ≤ 0.05) from two biological replicate experiments. a, Fold changes in gene expression at either 1, 4 or 18 h after activation compared to unactivated cells. Horizontal red dotted lines correspond to fold change threshold of ≥2. Numbers of genes upregulated or downregulated by ≥2× are indicated. b, Heatmaps represent expression levels of genes that were (1) differentially expressed by twofold at any time point with anti-IgM treatment and (2) associated with induced ATAC-seq peaks at either 1, 4 or 18 h time points as indicated above the heatmaps. Representative genes are shown on the right. c, Inducible transcripts were partitioned into three upregulated (patterns a–c) and three downregulated (patterns d–f) kinetic patterns by k-means clustering. Each graph represents the centroid profile of the average of z score of fold change (FC) compared with untreated cells at 1, 4 and 18 h. Total number of genes in each pattern are indicated on the right. d–f, RNA-seq analyses in activated B cells treated with IKK2 inhibitor BAY, or B cells that lack RelA or Rel. Differentially expressed genes were identified by DESeq2 (FDR ≤ 0.05). Genes that were induced at least twofold in control cells in each group were further separated into six kinetic patterns by k-means clustering. Graphs represent the centroid profile of the average row z score of fold change for genes in each cluster color coded as described below. Names of clusters and numbers of genes in each cluster are shown on top. The numbers of genes to which RelA (1 h) and Rel (18 h) were bound are indicated in red and blue font, respectively, below the patterns. Activation times are noted on the x axis. d, Kinetic patterns of BAY-responsive transcripts in activated C57BL/6 WT B cells in the absence (green) or presence of BAY (black). e, Kinetic patterns of differentially expressed transcripts in activated B cells from control RelAfl/fl (green) mice and RelA cKO mice (purple) activated with anti-IgM for times indicated on the x axis. f, Kinetic patterns of differentially expressed transcripts in activated B cells from control C57BL/6 mice (green) and Rel KO mice (orange) activated with anti-IgM for the indicated times. g, Representative browser tracks (mm9 annotation) from RNA-seq analyses of early (Egr2) and late (Axl) NF-κB target genes in WT and BAY-treated cells. Anti-IgM treatment times are shown on the right; transcriptional direction and transcription initiation sites are shown by red arrows.