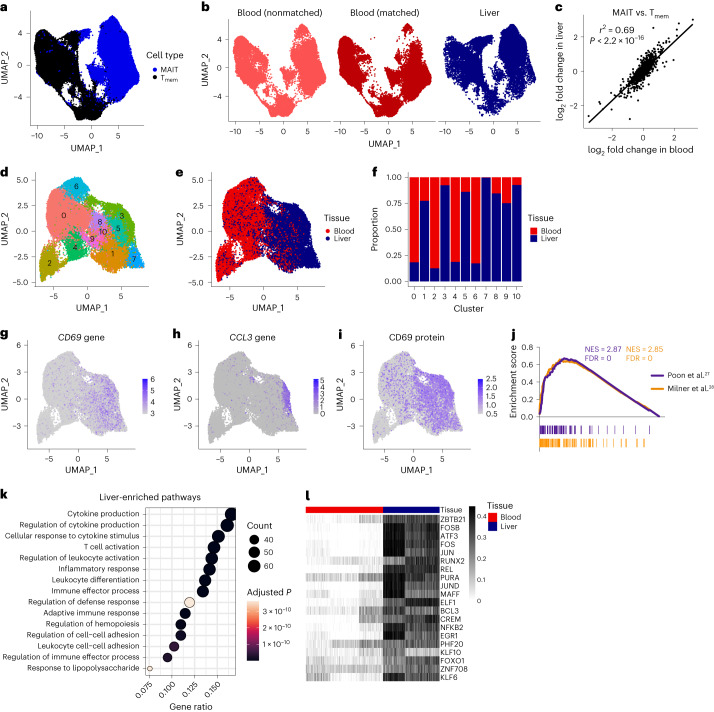
Fig. 1. Liver MAIT cells exhibit an activated, tissue-resident transcriptional and regulatory profile.
a, UMAP of blood and liver MAIT cells and conventional memory T (Tmem) cells colored by cell type. n = 89,456 cells from 12 donors. b, UMAP split by sample type, namely blood from healthy donors (nonmatched), blood from liver donors (matched) and liver. c, Pearson’s correlation between the log2 fold change in gene expression between MAIT and Tmem cells in the blood, and MAIT and Tmem cells in the liver. d,e, UMAP of matched blood and liver MAIT cells (n = 35,407 cells from six donors) colored by the 11 identified clusters (d) or by tissue (e). f, Proportion of cells in each cluster from the blood and liver. g–i, UMAPs colored by expression of CD69 (g) and CCL3 (h) genes or CD69 protein (i). j, Gene set enrichment analysis of liver compared with blood MAIT cells using published human and mouse tissue-resident memory T cell gene signatures. NES, normalized enrichment score. k, Over-representation analysis on the genes significantly upregulated in liver MAIT cells compared with blood MAIT cells. Top 15 gene ontology terms and associated Benjamini–Hochberg adjusted P values are shown. l, Heatmap showing activity (row-scaled AUCell scores) of the 20 most differentially active regulons (largest difference in average AUCell score) between matched blood and liver MAIT cells in Exp 1. n = 3 donors.