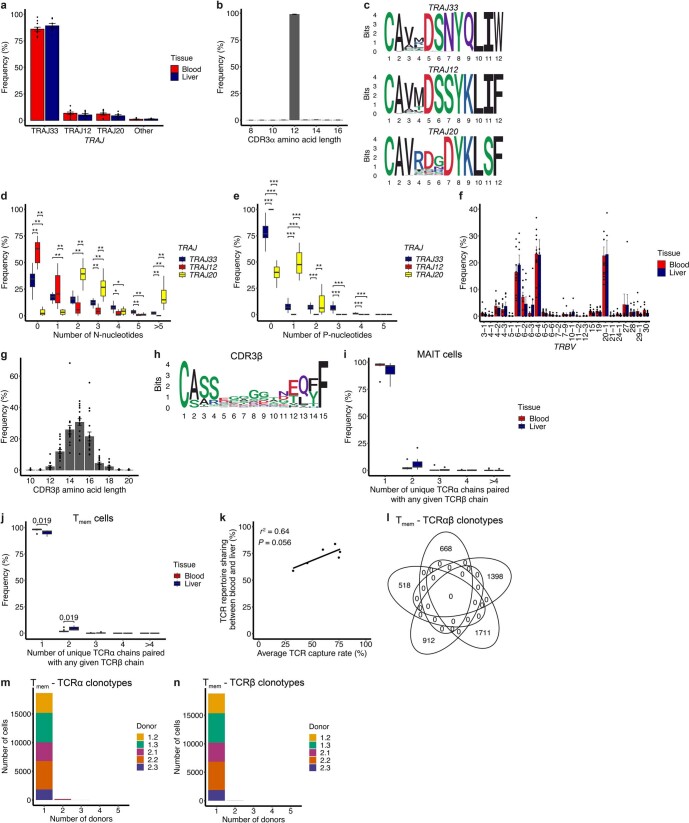
Extended Data Fig. 2. MAIT cells have a restricted TCRα but diverse TCRβ chain, resulting in private TCRαβ repertoires.
a, Proportion of blood and liver cells expressing TRAJ33, TRAJ12, TRAJ20 and other TRAJ gene segments. b, Distribution of CDR3α amino acid lengths. c, Sequence logos generated from all TRAJ33, TRAJ12 or TRAJ20 CDR3α amino acid sequences of length 12 (n = 26,529, 1,852 and 1,451 sequences, respectively). d,e, Frequency of N-nucleotides (d) and P-nucleotides (e) in TRAJ33, TRAJ12 and TRAJ20 TCRs from 12 donors. f, Proportion of blood and liver MAIT cells expressing different TRBV gene segments. Plot includes TRBV gene segments with a frequency >1% in at least one sample. g, Distribution of CDR3β amino acid lengths. h, Sequence logo generated from all MAIT cell CDR3β amino acid sequences of length 15 (n = 9,300 sequences). i,j, TCR chain pairing at the population level. Number of unique TCRα chains paired with any given TCRβ chain in blood and liver MAIT (i; n = 11 blood, 7 liver samples) or Tmem (j; n = 10 blood, 5 liver samples) cells. k, Pearson’s correlation between the average TCR capture rate (percentage of cells with a paired TRAV1-2 TCR) for a donor (n = 6) and percentage MAIT cell TCR repertoire sharing between matched blood and liver. l, Venn diagram showing the number of TCRαβ clonotypes shared between the five Tmem cell donors. m,n, Number of Tmem cells from each donor belonging to TCRα clonotypes (m) or TCRβ clonotypes (n) found in 1, 2, 3, 4, or 5 (all) donors. a-c and f-i show data from n = 18 samples (11 blood, 7 liver), 12 donors. Data in a, b, f, g are presented as mean ± s.e.m. In d, e, i, j, boxes span the 25th–75th percentiles, the midline denotes the median and whiskers extend to ± 1.5 × IQR. Points in i and j indicate outliers. Two-sided Wilcoxon rank-sum test (a, f, i, j) and two-sided Wilcoxon signed-rank test (d, e) for all pairwise comparisons. Benjamini-Hochberg adjusted P values are shown (nonsignificant results omitted). *P < 0.05, **P < 0.01, ***P < 0.001.