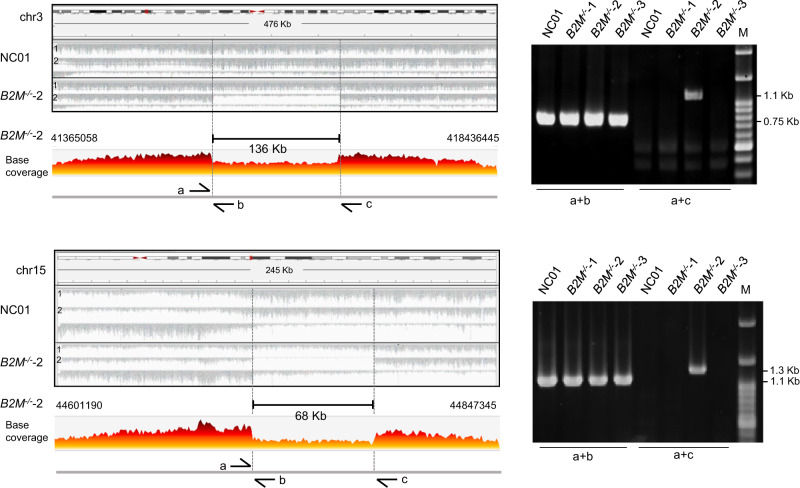
Fig. 3. PCR validation for the heterozygous large SVs in B2M-/-−2.
The phased BAM format was performed with the Integrative Genomics Viewer (IGV_2.7.2)48–50 sorted with 10x Genomics linked-read haplotype tag (HP; haplotype of the molecule that generated the read). The region of large SVs in the parental (NC01) and B2M-/-−2 can be phased into two strands with an HP tag (left panel). Agarose gel electrophoresis images of the PCR products amplified from NC01 genomic DNA and B2M knockouts (right panel). Primers a and b are designed to target the breakpoint junctions and primers a and b for the intact genomic region. The base coverage of large SVs in B2M-/-−2 was visualized by Loupe. M is the size marker. Primers are listed in Supplementary Table 12. n = 3 replicates for PCR analysis. Source data are provided as a Source Data file.