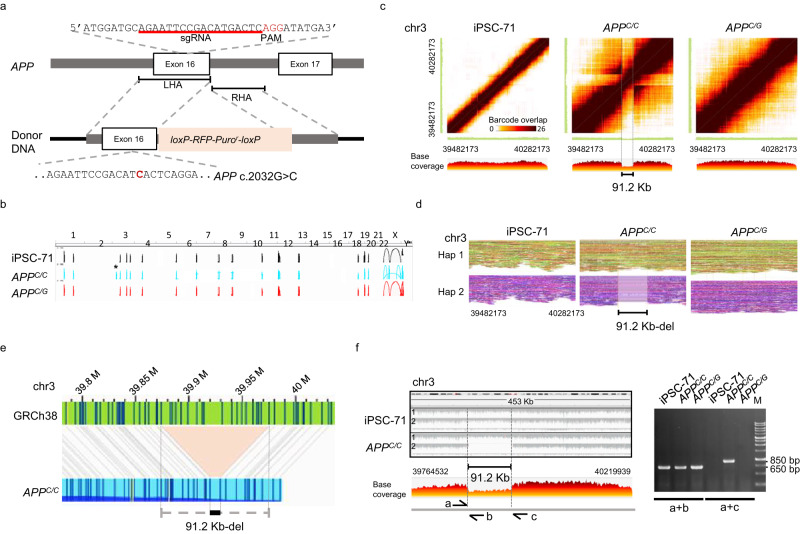
Fig. 4. Large SV was found after CRISPR-Cas9 mediated gene knock-in.
a The strategy for the base substitution of exon 16 of APP gene in human iPSC-71 by CRISPR-Cas9 mediated gene knock-in. The schematic presentation and the sequence of APP exon 16 loci with PAM, the targeting site for the sgRNA, and the donor DNA used for homology-directed repair (HDR) are shown. The loxP-RFP-Puror-loxP cassette was flanked by two homology arms. LHA, left homology arm (633 bp); RHA, right homology arm (582 bp). Boxes represent exons. b Large SV calls were constructed from linked-reads of the genome from iPSC-71, APPC/C, and APPC/G. The asterisk * indicates a large SV found in the APPC/C but not in the parental (iPSC-71) nor APPC/G. c Heat map of the overlapping barcodes in chr3:39482173–40282173 showing a heterozygous deletion in APPC/C. The matrix view was plotted by the Loupe, where the dark brown color represents the shared barcodes between the two genomic segments on the X- and Y-axis. The linear view represents the base coverage along the X-axis segment. d The phased reads graph indicates a 91.2 Kb-heterozygous deletion on chr3 in the APPC/C genome. Reads were partitioned into distinct haplotypes: haplotype1 (Hap1) and haplotype 2 (Hap2). e The optical genome mapping reveals a 91.2 Kb-heterozygous deletion (black bar) in chr3 of the APPC/C. The gray lines indicate the alignment between the reference (GRCh38; green) and assembled map (APPC/C; blue). The light red area shows the deletion. (f) PCR confirmation of the heterozygous deletion. PCR verification with primers a and b and using iPSC-71, APPC/C, and APPC/G genomic DNA as the template. The primer pair, a and b, were designed to amplify breakpoints of the deletion, which produce a unique amplicon in the mutant strand harboring the large SV; and the primer pair, a and b, were for the wild-type strand. The region of the large SV in the iPSC-71 and APPC/C was phased into two strands with an HP tag (left panel) and base coverage of APPC/C visualized by Loupe. M is the size marker. Primers are listed in Supplementary Table 12. n = 3 replicates for PCR analysis. Source data are provided as a Source Data file.