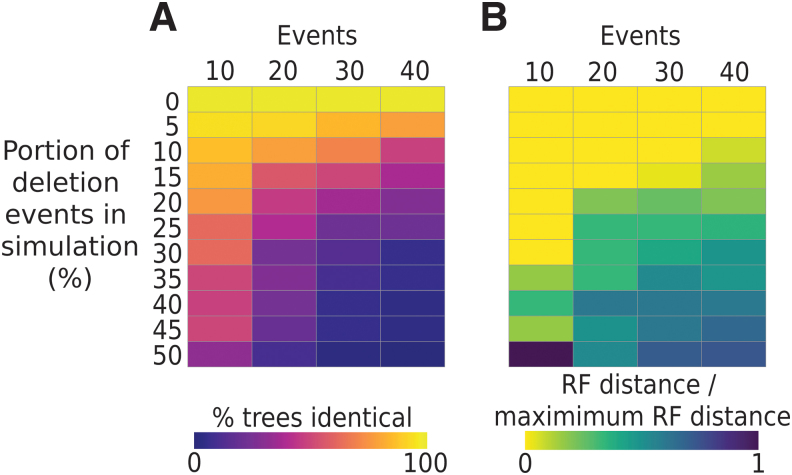
FIG. 6.
CRISPRtree can accurately recreate the true relationship between arrays when the frequency of spacer deletions is low. Simulated CRISPR arrays were produced using CCTK Evolve, and their relationships recorded in what will be referred to as the true tree topology. CRISPRtree was then used to analyze the simulated CRISPR arrays and to infer a maximum parsimony tree. The true topology of the simulated trees was compared with the topology inferred by CRISPRtree using RF distance (see the Methods section). To allow the calculation of summary statistics between multiple sets of trees, RF distance is presented as a proportion of the maximum theoretical distance for each pair of trees. Simulations were performed with a range of parameters: different proportions of spacer acquisition and deletion events (rows within each heatmap); and number of events for which each simulation was run (columns within each heatmap). For each set of parameters, 50 replicate simulations were run and two summary statistics are reported: (A) the proportion of trees produced by CRISPRtree that are identical to the true tree topology (RF distance of 0); (B) the median ratio of the observed RF distance to maximum theoretical RF distance between the CRISPRtree and true tree topology. RF, Robinson–Foulds.