Abstract
Efficient production of adeno-associated virus (AAV) vectors is a significant challenge. Human embryonic kidney HEK293T cells are widely used in good manufacturing practice facilities, producing higher yield of AAV vectors for clinical applications than HEK293 through the addition of a constitutive expression of SV40 large T antigen (SV40T), which stimulates Rep expression. However, the theoretical potential for tumorigenic consequences of a clinical AAV product containing residual DNA encoding SV40T, which may inhibit p53 growth suppressive functions is a safety concern. Although the risk is theoretical, to assure a low risk/high confidence of safety for clinical drug development, we have established a sensitive assay for assessment of functional full-length transcription competent SV40T DNA in HEK293T cell-produced AAV vectors. Using HEK293T generated 8, 9, and rh.10 serotype AAV vectors, the presence of SV40T in purified vector was assessed in vitro using quantitative polymerase chain reaction (qPCR) targeting a 129 bp amplicon combined with nested PCR targeting full-length SV40T DNA. Although low levels of the smaller amplicon were present in each AAV serotype, the full-length SV40T was undetectable. No transcription competent full-length SV40T DNA was observed by reverse transcription-quantitative polymerase chain reaction using an in vivo amplification of signal in mouse liver administered (2–10 × 1010 gc) 129 bp amplicon-positive AAV vectors. As a control for gene transfer, high levels of expressed transgene mRNAs were observed from each serotype AAV vector, yet, SV40T mRNA was undetectable. In vivo assessment of these three liver-tropic AAV serotypes, each with amplicon-positive qPCR SV40T DNA, demonstrated high transgene mRNA expression but no SV40T mRNA, that is, detection of small segments of SV40T DNA in 293T cell produced AAV inappropriately leads to the conclusion of residuals with the potential to express SV40T. This sensitive assay can be used to assess the level, if any, of SV40T antigen contaminating AAV vectors generated by HEK293T cells. ClinicalTrials.gov identifier: NCT03634007; NCT05302271; NCT01414985; NCT01161576.
Keywords: AAV, gene therapy, clinical, GMP production
INTRODUCTION
Adeno-associated virus (AAV) is one of the most commonly used gene transfer vectors used in gene therapy.1–4 Based on the encouraging therapeutic efficacy and safety profile in clinical trials, several AAV vectors have progressed to commercialization. One of the commonly used, scalable, good manufacturing practice (GMP) AAV vector production systems involves plasmid transfection mediated production in human embryonic kidney HEK293 or HEK293T cells.5–10 The difference in the two cell lines is that while HEK293 and HEK293T cells are transformed to express adenovirus E1A and E1B genes, the HEK293T cells also include the SV40 large T antigen (SV40T). Large T antigen provides enhanced helper function for AAV production by stimulating Rep expression.11 For AAV production, compared to HEK293 cells, HEK293T packaging cells grow faster and yield 5–11 times more AAV resulting in significant reduction in the cost of vector production.12
Despite these advantages, regulatory agencies have expressed concern for the contamination in AAV preparations of the sequence for the SV40T in HEK293T cells based on a theoretical safety concern with respect to SV40T-induced tumorigenesis.13–15 In the production of AAV vectors, DNase treatment digests nonencapsidated residual host cell DNA from the product, however, encapsidated host cell DNA is protected from digestion by the AAV capsid, which is indistinguishable from the desired AAV product and therefore can persist through purification to be present in the final product.16,17 As a result, it is possible that there is SV40T DNA in the AAV drug, inseparable from the transgene encapsidated AAV. Despite numerous studies finding no evidence for this SV40T-induced tumorigenicity in humans,18–22 it is incumbent on the sponsor of an AAV gene therapeutic to demonstrate robust safety before clinical trial.
To address the issue of possible SV40T antigen DNA in AAV vectors generated by HEK293T cells, we have generated a highly sensitive method to quantify SV40T in final GMP AAV products produced in HEK293T cells. We assessed the presence of full-length transcription-competent SV40T DNA in 293T cell-produced AAV vectors using AAVrh.10, AAV9, and AAV8 vectors encoding various transgenes. The presence of SV40T in purified vector was assessed by directly analyzing the purified AAV vector by quantitative polymerase chain reaction (qPCR) targeting the entire SV40T sequence, amplified by nested PCR (nPCR). As a further measure of transcription competent full-length SV40T in AAV preparations, we assessed each of the AAV vectors for SV40T mRNA expression in liver in mice following high-dose intravenous administration.
METHODS
AAV vectors
The AAV vector DNA consists, from 5′ to 3′, the AAV2 5′ inverted terminal repeat (ITR) containing the packaging signal, which directs the encapsidation of the recombinant AAV genome, CAG promoter consisting of CMV enhancer, chicken β-actin promoter and splice donor and rabbit β-globin intron with splice acceptor, the transgene cDNA, rabbit β-globin polyA signal, and the AAV2 3′ ITR. The transgene expression cassettes were packaged into AAVrh.10, AAV9, or AAV8 capsids.
The AAVrh.10 vectors were produced as described previously.23 In brief, the expression plasmids pAAVmCherry encoding mCherry cDNA or pAAVhFXN encoding human frataxin or pAAVhGalc encoding human galactosylceramidase (600 μg) and the AAVrh.10 capsid-Ad5 helper hybrid plasmid pPAKMArh.10 (1.2 mg) were cotransfected into 40 × 15 cm dishes of HEK293T cells containing an integrated copy of the adenovirus E1 gene using polyethyleneimine (PEI) transfection reagent (Polysciences, Warrington, PA, USA). The AAV9mCherry vector was produced by cotransfection of HEK293T cells with pAAVmCherry encoding mCherry cDNA (600 μg), AAV9 packaging plasmid (600 μg) and Ad helper plasmid ΔF6 (1.2 mg) using PEI transfection reagent. The AAV8hAAT-AVL vector was produced by cotransfection of HEK293T cells with pAAVAAT-AVL encoding oxidation-resistant human αl-antitrypsin cDNA (55 μg),24 AAV8 capsid plasmid (550 μg) and Ad5 helper plasmid ΔF6 (1.1 mg) using PEI transfection reagent.
At 72 h post-transfection, cells were harvested and crude viral lysate (CVL) prepared by 5 × freeze/thaw cycles followed by centrifugation at 3,000 g at 4°C for 20 min and the supernatant was collected and treated with benzonase (100 U/mL). The AAVrh.10 vectors were then purified from the supernatant by 15–54% discontinuous iodixanol gradient centrifugation and high performance anion exchange column (QHP) chromatography at pH 9.0 (GE Healthcare, Piscataway, NJ, USA). The AAV9 vector was purified from the CVL by the same iodixanol gradient centrifugation and QHP anion exchange column chromatography at pH 7.5.23 The AAV8 vector was purified from the CVL by iodixanol gradient centrifugation and vector band was collected as pure vector without the ion exchange chromatography.25 All purified vectors were concentrated using BioMax 100K membrane concentrators (Millipore, Billerica, MA, USA) and stored in phosphate-buffered saline, pH 7.4 at −80°C.
Vector genome titers of all AAV serotype vectors were determined by TaqMan real-time PCR (qPCR) using a CMV-chicken ß-actin promoter (CAG)-specific primer–probe set (forward primer: 5′-GTCAATGGGTGGAGTATTTACGG-3′, reverse primer: 5′-AGGTCATGTA CTGGGCATAATGC-3′ and probe: FAM-CAAGTGTATCATATGCCAAGTACGCCCCC-TAMRA), designed using Primer Express software (Applied Biosystems, Foster City, CA, USA). Purified vectors were digested with proteinase K in the presence of 0.5% sodium dodecyl sulfate (SDS) and 25 mM ethylenediaminetetraacetate (EDTA) at 55°C for 1 h followed by inactivation of the protease at 95°C for 15 min. The DNA released in the digest was then used as a template for qPCR using pAAV-CAG-hCLN2 plasmid DNA encoding human CLN2 cDNA under the CAG promoter as reference standard.
In vitro assessment of SV40T DNA in purified AAV vectors
SV40T amplicon by qPCR
AAV vectors (1.9 × 1010 to 1.8 × 1011 gc) were treated with proteinase K in presence of 0.5% SDS and 25 mM EDTA at 55°C, 1 h followed by 95°C, 15 min. The vector-associated DNA was purified by phenol:chloroform:isoamyl alcohol (24:1:1) extraction and isopropanol (75%) precipitation. The precipitated DNA pellet was washed twice with 70% ethanol, resuspended in water, and stored at −20°C. Vector genomic DNA was isolated from the purified AAV vectors and quantified by qPCR using CAG-specific primers and probe. The SV40T DNA was then assessed by qPCR using the forward primer 5′-ATGCTCATCAACCTGACTTTGGA-3′, reverse primer 5′-GGCCATTGTTGCAGTACATTG-3′, and probe FAM-TCTGGATGCAACTGA-NFQ. A standard curve was generated with SV40T cDNA plasmid spanning the range of 101–108 copies.
Full-length SV40T DNA by nPCR
The DNA isolated from purified AAV was first PCR amplified using 5′-terminal forward primer 5′-ATGGATAAAGTTTTAAACAGAGAGGAATCTTTGCAGC-3′ and 3′-terminal reverse primer 5′-TTATGTTTCAGGTTCAGGGGGAGGTGTGGGAGG-3′ under the following conditions: 94°C for 5 min, 35 cycles at 94°C for 30 s—55°C for 30 s—68°C for 150 s, followed by 68°C for 7 min and finally held at 4°C. The full-length SV40T-specific PCR product was then further amplified by second PCR with internal forward primer 5′-ATGGAACTGATGAATGGGAGCAGTGGTGGAATGCC-3′ and internal reverse primer 5′-TGGCCTGCAGTGTTTTAGGCACACTG TACTCATTCATG-3′ under the following conditions: 94°C for 5 min, 25 cycles at 94°C for 30 s—55°C for 30 s—68°C for 90 s, followed by 68°C for 7 min and finally held at 4°C. The PCR products were analyzed by 1% agarose gel electrophoresis and ethidium bromide staining.
Vector administration and preparation of liver homogenates
AAV vectors were administered in 100 μL total volume in phosphate-buffered saline (PBS) to 6–8 week old C57Bl/6 male mice (n = 4 or 5) via intravenous route (tail vein). At 2 days postadministration, mice were euthanized by CO2 asphyxiation and immediately perfused by cardiac administration of 40 mL cold PBS, then livers were harvested. The livers were homogenized as previously described.26 In brief, whole liver was homogenized in 1.5 mL lysis buffer (10 mM HEPES-KOH, pH 7.4, 5 mM mannitol, and 1% Triton X-100 in water) using Tissue Lyser LT (Qiagen, Valencia, CA, USA) for 2 × 10 min at oscillation of 50/s. The homogenate was centrifuged at 10,000 g for 5 min and supernatant was collected and stored in 200 μL aliquots at −80°C.
Murine liver DNA and RNA isolation
Genomic DNA was isolated from AAV vector-administered mice liver homogenate using a DNeasy kit (Qiagen). In brief, 200 μL liver homogenate was incubated with 20 μL proteinase K and 4 μL RNase A (100 mg/mL) at 23°C for 2 min followed by incubation in buffer at 56°C for 10 min. The digest was mixed with ethanol and was loaded onto a DNeasy column. The column was centrifuged at 6,000 g for 30 s at room temperature, was washed with buffer AW1 followed by AW2 and finally DNA was eluted in 100 μL elution buffer.
Total RNA was isolated from AAV vector-administered mice liver homogenate using RNeasy kit according to the manufacturer's protocol (Qiagen). In brief, liver homogenate (200 μL) was mixed with 400 μL RLT buffer [from test kit] followed by 600 μL 70% ethanol and loaded onto a RNeasy column. The column was centrifuged at 8,000 g for 15 s and washed with buffer RW1 (from test kit). Then 25 U of RNase-free DNase I in RDD buffer (from test kit) was added to the column and incubated at room temperature for 15 min. The column was washed with buffer RW1 followed by buffer RPE and finally RNA was eluted with 35 μL of RNase free water. The RNA was quantified by absorbance at 260 nm using Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and stored at −80°C.
A preparation of RNA using the identical process was subjected to RIN analysis and resulted in good quality outcome with an average 8.5 and standard deviation of 0.7 providing qualification of the method.
Assessment of vector genome and transgene mRNA levels in liver
AAV vector genome level in liver homogenate was assessed by qPCR using CMV-specific primers and probe. The transgene mRNAs were assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using transgene-specific primers and probes. To synthesize cDNA, reverse transcription of total RNA was carried out following the procedure as described previously.26 The mCherry cDNA level was assessed using the forward primer 5′-CACCTACAAGGCCAAGAAGC-3′, reverse primer 5′-TGTCCAGCTTGATGTTGACG-3′ and probe FAM-CAGCTGCCCGGCGCCTACAA-NFQ. The hFXN cDNA was assessed using the primers and probes as described previously.26 Galc cDNA was assessed using human GalC-specific primers and probe (Thermo Fisher Scientific). hAAT-AVL was assessed using forward primer 5′-CCCGCCTGAGGTGAAGTT-3′, reverse primer 5′-GGTGACTTCGTATTCTGTTCGATCA-3′ and probe FAM-CACGAACGGCTTATTG-NFQ. Transgene mRNA specific RT-qPCR reactions used corresponding cDNA encoding plasmids as reference standards.
SV40T mRNA levels in liver
The SV40T mRNA level in AAV vector-administered liver homogenate was assessed by RT-qPCR. Total cDNA was synthesized by reverse transcription (Thermo Scientific, Somerset, NJ, USA) using 1 μg total RNA and the cDNA (200 ng) was used for qPCR using SV40T-specific primers and probe. The cDNA synthesized from naive mice liver served as the negative control. A plasmid containing SV40T cDNA was used as reference standard. The SV40T cDNA detection limit of the qPCR was determined by spiking known amounts of SV40T cDNA into naive liver cDNA (200 ng).
Results
Presence of SV40T DNA in purified AAV vectors
To assess residual SV40T DNA in AAV vectors, five vectors were produced in HEK293T cells, AAVrh.10mCherry, AAVrh.10FXN, AAVrh.10GALC, AAV9mCherry, and AAV8AAT-AVL. The vectors were purified as described in Methods section. The purity for each was evaluated by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining. Three capsid proteins, VP1, VP2, and VP3, and no cellular proteins were seen indicating high purity of all vectors. The total DNA isolated from at least 1011 gc purified vector was followed by qPCR analysis with SV40T-specific primers and probe encompassing a 129 bp amplicon, which targeted 1,629–1,758 bp of the 2,473 bp full length SV40T cDNA. In vitro assessment of these vectors using a standard assay of a small target sequence by qPCR showed the levels 3.2, 7.9, 8.7, 77.4, and 342.7 copies of SV40T DNA in 1010 AAV for AAVrh.10mCherry, AAVrh10FXN, AAVrh.10Galc, AAV9mCherry, and AAV8AAT-AVL, respectively (Table 1).
Table 1.
SV40T DNA in different liver-tropic adeno-associated virus serotype vectors
| Vector (kb) | In Vitro |
In Vivo |
|||||
|---|---|---|---|---|---|---|---|
| Vector DNA Analyzed (gc)a | SV40T Amplicon/1010 AAV Genome (Range)b | SV40T Full Length/1010 AAV Genome Rangec | Vector Dose (gc/kg)d | Vector DNA Copies/200 ng (Range)e | Transgene mRNA Copies/200 ng (Range)f | SV40T mRNA Copies/200 ng (Range)g | |
| AAVrh.10mCherry (3.2) | 1.8 × 1011 | 3.2 (2.5–3.9) | UD | 5 × 1012 | 5.1 × 105 (4.1 × 105–6.3 × 105) | 8.2 × 104 (3.4 × 104–2.0 × 105) | 38.1 (29.8–48.6), p > 0.4 |
| AAVrh.10hFXN (3.0) | 9.8 × 1010 | 7.9 (5.7–10) | UD | 5 × 1012 | 9.3 × 105 (3.6 × 105–1.9 × 106) | 1.3 × 103 (8.8 × 102–3.2 × 103) | 145.3 (98.2–214.9), p > 0.7 |
| AAVrh.10Galc (4.2) | 1.5 × 1011 | 8.7 (7.6–9.8) | UD | 5 × 1012 | 4.4 × 105 (3.8 × 105–5.6 × 105) | 1.8 × 103 (1.3 × 103–4.8 × 103) | 70.4 (43.9–112.9), p > 0.4 |
| AAV9mCherry (3.2) | 1.9 × 1010 | 77.4 (65.2–89.5) | UD | 5 × 1012 | 2.5 × 106 (1.4 × 106–3.1 × 106) | 2.5 × 104 (6.1 × 103–5.7 × 104) | UD |
| AAV8AAT-AVL (3.7) | 6.2 × 1010 | 342.7 (247.3–438.1) | UD | 5 × 1012 | 3.9 × 104 (2.5 × 104–9.2 × 104) | 1.7 × 104 (7.6 × 103–3.7 × 104) | 29.9 (3.9–229.4), p > 0.4 |
SV40T DNA was assessed in AAV vectors using 1.9 × 1010–1.8 × 1011 genome copies by qPCR; the SV40T DNA was expressed as copies/1010 AAV genome2.
SV40T DNA amplicon assessed by qPCR1 producing 129 bp spanning 1,629–1,758 bp of the SV40T DNA
SV40T DNA full length copies were assessed by nPCR.
AAV vectors (5 × 1012 gc/kg) were administered to C57BL/6 male mice (n = 4/5) via the intravenous route.
AAV vector DNA in liver was assessed by qPCR using 200 ng total DNA/assay.
Transgene mRNA copies in liver were assessed by RT-qPCR using mRNA-specific primers-probe and 200 ng total RNA/assay.
SV40T mRNA copies in liver were assessed by RT-qPCR using SV40T-specific primers-probe and 200 ng total RNA/assay p values compared to naive (n = 19) level 90.3 (28.5–286.6).
AAV, adeno-associated virus; nPCR, nested PCR; qPCR, quantitative PCR; RT-qPCR, reverse transcription-qPCR; UD, undetected (limit of detection 10 copies).
Full-length SV40T DNA undetectable in purified AAV vectors
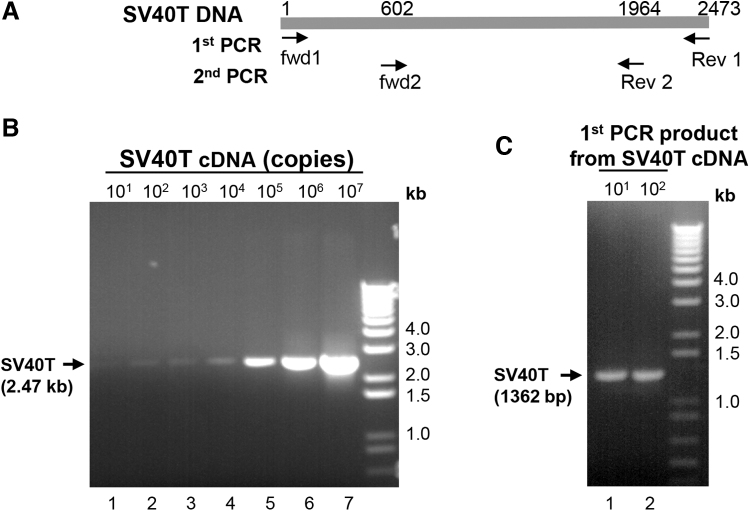
nPCR provided a quantitative measure of transcription competent full length SV40T DNA in total DNA isolated from the purified AAV vectors. Purified vector DNA (1.8 × 1011 genome copies) was PCR amplified using 3′ terminal forward and 5′ terminal reverse primers. For quantification, PCR amplified full length SV40T DNA products from 101 to 107 copies were used as reference standards in gel electrophoretic analysis. For each AAV vector, no full-length SV40T DNA was detected with the assay limit of detection of 100 copies in SV40T cDNA (Fig. 1 and Table 1).
Figure 1.
Assessment of full-length SV40T DNA in AAVrh.10 vector by nPCR. The DNA (1.8 × 1011 gc) isolated from purified AAVrh.10mCherry was used in the SV40T-specific PCR. (A) Schematic of PCR primers annealing positions on the SV40T cDNA. (B) First PCR using SV40T DNA terminal primers. Following PCR, the DNA was resolved in 1% Agarose gel and stained with EtBr. PCR products from 101 to 107 copies SV40T DNA was used as standard. (C) Second PCR using SV40T internal primers. The DNA product of the first PCR was further amplified by nPCR using SV40T internal primers. The DNA from first PCR of 101 and 102 copies SV40T DNA were used as standard. The PCR products were resolved in 1% agarose gel and stained with EtBr. EtBr, ethidium bromide; nPCR, nested PCR.
To increase the sensitivity of the assay, the PCR product obtained from the vector DNA was further amplified by nPCR using SV40T DNA-specific internal forward and reverse primers. In parallel, the full-length PCR products from SV40T DNA of 101–102 copies were amplified by nPCR. No SV40T DNA nPCR product was observed from the DNA of any AAV sample in an assay with a sensitivity of 10 copies of SV40T DNA (Fig. 1 and Table 1).
DNA from hepatotropic AAV vectors produced in 293T cells does not express SV40T mRNA in vivo
Transcription of AAV-derived DNA to mRNA in the mouse liver provides signal amplification of full-length functional DNA genes; assaying for SV40T mRNA provides a sensitive indicator for the presence of functional SV40T DNA. A high dose of AAV vectors (5 × 1012 genome copies/kg) was administered intravenously for each vector to mice and delivery of each was confirmed via the detection of high levels of vector genomic DNA (depending on AAV serotype and transgene ranged from 3.9 × 104 to 2.5 × 106 copies/200 ng total DNA) observed in the liver 2 days postvector administration (Table 1). High levels of the delivered AAV vector-expressed transgene in the liver were found by RT-qPCR (transgene mRNA levels were 1.3 × 103 to 4.1 × 105 copies/200 ng total RNA, Table 1).
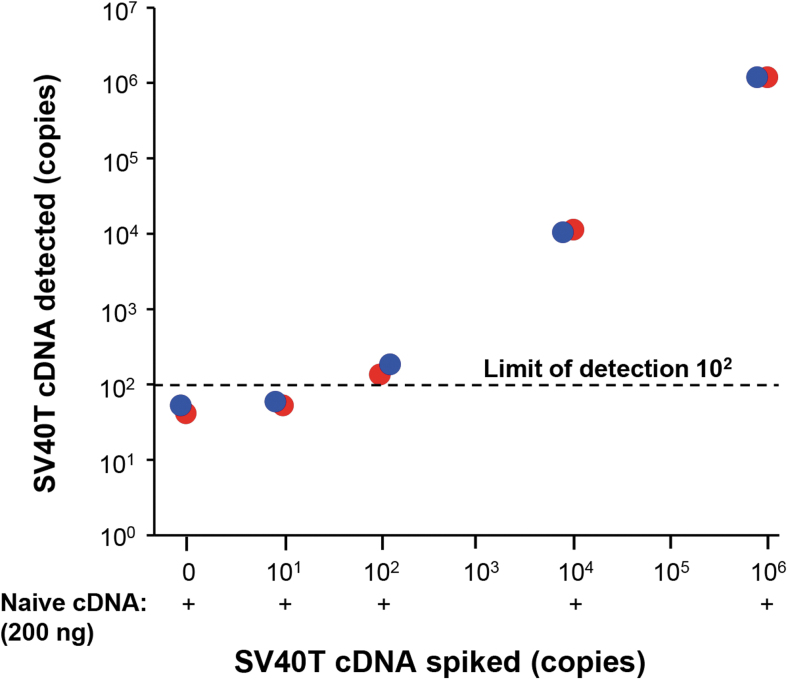
Finally, we assessed liver-amplified SV40T mRNA from potentially present transcription competent full-length SV40T DNA impurity in each AAV vector. To identify the sensitivity of the method, SV40T cDNA was spiked at increasing amounts into naive liver total cDNA and subsequent RT-qPCR analysis indicated the limit of detection was 100 copies (Fig. 2). SV40T mRNA expression in each of the AAV-administered mice liver was assayed by RT-qPCR using SV40T-specific primers for the 129 nt amplicon. For context, the level of SV40T mRNA copies in naive mouse liver was similarly assessed (n = 19) and levels averaged 90.3 (range 28.5–286.6). No significant differences were found between SV40T mRNA levels between any of the AAV vectors administered (n = 4/5) and naive mice (n = 19, p > 0.4, Table 1).
Figure 2.
Determination of the limit of detection in assessment of SV40T mRNA expression following intravenous administration of an AAVrh.10 vector. AAVrh.10mCherry (1011 gc) was administered to 6–8-week old C57BL/6 male mice (n = 4) via tail vein. After 2 days, SV40T mRNA expression was assessed by measuring the mRNA level in liver. For comparison, SV40T cDNA was spiked into naive liver cDNA. To determine the limit of detection, naive liver cDNA (200 ng) was spiked with 101 to 106 copies of SV40T cDNA and the spiked cDNA was quantified by qPCR using SV40T primers-probe. Technical replicates are shown in red and blue symbols.
Therefore, despite the apparent measured quantity by qPCR of a small target sequence for the SV40T in packaged AAV vectors, no measurable productive transcription of SV40 Large T antigen mRNA is observed even after in vivo signal amplification.
DISCUSSION
AAV vectors are being widely used for therapeutic gene delivery and there is an increased demand for high-yield vector production. In this regard, the HEK293T cell line has the advantage of producing higher yield AAV vectors with lower costs due to the rapid rate of cell growth.27 All production methods require assessment of residual impurities and since HEK293T cells have the SV40T sequence in the cell genome, there is the theoretical concern for the tumorigenic potential of residual SV40 large T antigen DNA (SV40T). Despite significant evidence for the lack of such SV40T untoward effects, including studies of recipients of the SV40-contaminated early polio vaccine administered to millions,28 this concern requires additional data to minimize perceived risk.
In this study, we have developed a sensitive method using in vitro and in vivo assessments of SV40T DNA in purified AAV vectors produced in HEK293T cells with multiple AAV serotypes and transgenes. Although standard methods to assay residual SV40T DNA use qPCR analysis targeting a short DNA segment was positive for this contaminant in purified AAV vectors, full length copies were not detected by using nPCR with a sensitivity of 10 copies. Finally, as a measure of transcription competent full-length SV40T DNA, we assessed mRNA expression by amplifying the signal in the target organ liver of mice following intravenous administration of each of five AAV vectors. The qPCR and RT-qPCR analyses of the liver homogenate demonstrated high levels of both vector DNA and transgene mRNA expression confirming transduction and transgene expression. In contrast, SV40T-specific RT-qPCR showed no SV40T mRNA expression.
Thus, despite the presence of small segments of SV40T DNA typically used to evaluate residual contaminants in a purified AAV preparation, there was no measurable full length SV40T DNA nor SV40T mRNA in an in vivo assay designed to maximize signal using liver for amplification.
AAV vectors produced in HEK293T cells require, as in all production methods, rigorous downstream purification and quality control to reduce DNA impurities. But these steps can only remove impurities external to the encapsidated AAV payload.29–31 DNA within the AAV particle are DNase resistant and impossible to remove, degrade or isolate from the desired AAV product.7,32,33 Recombinant AAV vectors have been shown to nonspecifically package plasmid-derived rep/cap DNA and packaging cell-derived adenovirus sequences.8,34 In this regard, there has been concern for AAV vectors produced in HEK293T cell system to package SV40T DNA.8 These packaged DNAs present perceived or theoretical safety concerns in the context that other studies have shown sequences derived from the rep-cap plasmid to be present at a level of 0.02–1% of vector genome.34
While the packaged nontransgene DNA has been shown to be heterogeneous in size, a functional outcome has been the exception. One example is the incorporation of transcriptionally active AAV cap DNA in AAV2, 7, and 8 vectors at levels ranging from 0.4% to 1%,29,35 with a possible outcome of induced cytotoxic T lymphocyte-mediated lysis of cells expressing these capsid proteins.36,37 But considering that moles of plasmid used in production is typically 100-fold greater than the number of packaging host cell genomes suggests that incorporation of a particular host cell genome DNA into the AAV is much less likely than the DNA from the plasmids used in production.
Further, the likelihood of the delivery of any unintended functional DNA is linked to the dose of vector administered, suggesting that studies with very large dosing regimens should have higher stringency for limits of residual non-transgene DNA. This should include packaging of adenovirus E1 DNA from HEK293 as well as HEK293T cells into the AAV vectors, for which several studies have raised a safety concern.15,38 Packaging of these adenovirus DNA sequences in HEK293 cell-produced AAV vector was investigated using next-generation single-stranded virus sequencing method and no detectable level of DNA was found in purified vector.16 In summary, we have developed a sensitive assay method using a combination of in vitro and in vivo assays of HEK293T-produced AAV vectors, irrespective of serotype, with qPCR detectable levels of SV40T DNA that demonstrate no measurable SV40 large T antigen mRNA, providing support for the safe clinical use of these vectors.
ACKNOWLEDGMENTS
We thank Alvin Chen, Herg Zhang, Michael Kuckyr, Humam Al Rubaye for technical work and N. Mohamed for editorial assistance.
AUTHORs' CONTRIBUTIONS
S.M.K. developed the experimental strategy; S.M.K., R.G.C., B.P.D., S.C., and H.L. designed the experiments; B.P.D. and J.B.R. performed the experiments; S.M.K. and B.P.D. analyzed the data; B.P.S., S.M.K., D.S., and R.G.C. wrote the article.
AUTHOR DISCLOSURE
No competing financial interests exist.
FUNDING INFORMATION
These studies were supported by R61HL151355 and the Department of Genetic Medicine.
REFERENCES
- 1. Coura Rdos S, Nardi NB. The state of the art of adeno-associated virus-based vectors in gene therapy. Virol J 2007;4:99; doi: 10.1186/1743-422x-4-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Naso MF, Tomkowicz B, Perry WL, III, et al. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 2017;31(4):317–334; doi: 10.1007/s40259-017-0234-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goswami R, Subramanian G, Silayeva L, et al. Gene therapy leaves a vicious cycle. Front Oncol 2019;9:297; doi: 10.3389/fonc.2019.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Au HKE, Isalan M, Mielcarek M. Gene therapy advances: A meta-analysis of AAV usage in clinical settings. Front Med (Lausanne) 2021;8:809118; doi: 10.3389/fmed.2021.809118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zolotukhin S, Byrne BJ, Mason E, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 1999;6(6):973–985; doi: 10.1038/sj.gt.3300938 [DOI] [PubMed] [Google Scholar]
- 6. Grimm D. Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods 2002;28(2):146–157; doi: 10.1016/s1046-2023(02)00219-0 [DOI] [PubMed] [Google Scholar]
- 7. Chadeuf G, Ciron C, Moullier P, et al. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol Ther 2005;12(4):744–753; doi: 10.1016/j.ymthe.2005.06.003 [DOI] [PubMed] [Google Scholar]
- 8. Allay JA, Sleep S, Long S, et al. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum Gene Ther 2011;22(5):595–604; doi: 10.1089/hum.2010.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schnödt M, Büning H. Improving the quality of adeno-associated viral vector preparations: the challenge of product-related impurities. Hum Gene Ther Methods 2017;28(3):101–108; doi: 10.1089/hgtb.2016.188 [DOI] [PubMed] [Google Scholar]
- 10. Kimura T, Ferran B, Tsukahara Y, et al. Production of adeno-associated virus vectors for in vitro and in vivo applications. Sci Rep 2019;9(1):13601; doi: 10.1038/s41598-019-49624-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogston P, Raj K, Beard P. Productive replication of adeno-associated virus can occur in human papillomavirus type 16 (HPV-16) episome-containing keratinocytes and is augmented by the HPV-16 E2 protein. J Virol 2000;74(8):3494–3504; doi: 10.1128/jvi.74.8.3494-3504.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park JY, Lim BP, Lee K, et al. Scalable production of adeno-associated virus type 2 vectors via suspension transfection. Biotechnol Bioeng 2006;94(3):416–430; doi: 10.1002/bit.20776 [DOI] [PubMed] [Google Scholar]
- 13. Sullivan CS, Pipas JM. T antigens of simian virus 40: Molecular chaperones for viral replication and tumorigenesis. Microbiol Mol Biol Rev 2002;66(2):179–202; doi: 10.1128/mmbr.66.2.179-202.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005;24(52):7729–7745; doi: 10.1038/sj.onc.1209046 [DOI] [PubMed] [Google Scholar]
- 15. Baldo A, van den Akker E, Bergmans HE, et al. General considerations on the biosafety of virus-derived vectors used in gene therapy and vaccination. Curr Gene Ther 2013;13(6):385–394; doi: 10.2174/15665232113136660005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lecomte E, Tournaire B, Cogné B, et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol Ther Nucleic Acids 2015;4(10):e260; doi: 10.1038/mtna.2015.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tai PWL, Xie J, Fong K, et al. Adeno-associated virus genome population sequencing achieves full vector genome resolution and reveals human-vector chimeras. Mol Ther Methods Clin Dev 2018;9:130–141; doi: 10.1016/j.omtm.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Worgall S, Sondhi D, Hackett NR, et al. Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum Gene Ther 2008;19(5):463–474; doi: 10.1089/hum.2008.022 [DOI] [PubMed] [Google Scholar]
- 19. Nathwani AC, Reiss UM, Tuddenham EG, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 2014;371(21):1994–2004; doi: 10.1056/NEJMoa1407309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tardieu M, Zérah M, Husson B, et al. Intracerebral administration of adeno-associated viral vector serotype rh.10 carrying human SGSH and SUMF1 cDNAs in children with mucopolysaccharidosis type IIIA disease: Results of a phase I/II trial. Hum Gene Ther 2014;25(6):506–516; doi: 10.1089/hum.2013.238 [DOI] [PubMed] [Google Scholar]
- 21. Clément N, Grieger JC. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol Ther Methods Clin Dev 2016;3:16002; doi: 10.1038/mtm.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sondhi D, Kaminsky SM, Hackett NR, et al. Slowing late infantile Batten disease by direct brain parenchymal administration of a rh.10 adeno-associated virus expressing CLN2. Sci Transl Med 2020;12(572):eabb5412; doi: 10.1126/scitranslmed.abb5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De BP, Heguy A, Hackett NR, et al. High levels of persistent expression of alpha1-antitrypsin mediated by the nonhuman primate serotype rh.10 adeno-associated virus despite preexisting immunity to common human adeno-associated viruses. Mol Ther 2006;13(1):67–76; doi: 10.1016/j.ymthe.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 24. Sosulski ML, Stiles KM, Frenk EZ, et al. Gene therapy for alpha 1-antitrypsin deficiency with an oxidant-resistant human alpha 1-antitrypsin. JCI Insight 2020;5(15):e135951; doi: 10.1172/jci.insight.135951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pignataro D, Sucunza D, Vanrell L, et al. Adeno-associated viral vectors serotype 8 for cell-specific delivery of therapeutic genes in the Central Nervous System. Front Neuroanat 2017;11:2; doi: 10.3389/fnana.2017.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De BP, Chen A, Salami CO, et al. In vivo potency assay for adeno-associated virus-based gene therapy vectors using AAVrh.10 as an example. Hum Gene Ther Methods 2018;29(3):146–155; doi: 10.1089/hgtb.2017.246 [DOI] [PubMed] [Google Scholar]
- 27. Bae DH, Marino M, Iaffaldano B, et al. Design and testing of vector-producing HEK293T cells bearing a genomic deletion of the SV40 T antigen coding region. Mol Ther Methods Clin Dev 2020;18:631–638; doi: 10.1016/j.omtm.2020.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strickler HD, Rosenberg PS, Devesa SS, et al. Contamination of poliovirus vaccines with simian virus 40 (1955–1963) and subsequent cancer rates. JAMA 1998;279(4):292–295; doi: 10.1001/jama.279.4.292 [DOI] [PubMed] [Google Scholar]
- 29. Hauck B, Murphy SL, Smith PH, et al. Undetectable transcription of cap in a clinical AAV vector: Implications for preformed capsid in immune responses. Mol Ther 2009;17(1):144–152; doi: 10.1038/mt.2008.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wright JF. Product-related impurities in clinical-grade recombinant AAV vectors: Characterization and risk assessment. Biomedicines 2014;2(1):80–97; doi: 10.3390/biomedicines2010080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mietzsch M, Hering H, Hammer EM, et al. OneBac 2.0: Sf9 cell lines for production of AAV1, AAV2, and aav8 vectors with minimal encapsidation of foreign DNA. Hum Gene Ther Methods 2017;28(1):15–22; doi: 10.1089/hgtb.2016.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nony P, Chadeuf G, Tessier J, et al. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: A model for generation of rep-positive AAV particles. J Virol 2003;77(1):776–781; doi: 10.1128/jvi.77.1.776-781.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Halbert CL, Metzger MJ, Lam SL, et al. Capsid-expressing DNA in AAV vectors and its elimination by use of an oversize capsid gene for vector production. Gene Ther 2011;18(4):411–417; doi: 10.1038/gt.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Penaud-Budloo M, François A, Clément N, et al. Pharmacology of recombinant adeno-associated virus production. Mol Ther Methods Clin Dev 2018;8:166–180; doi: 10.1016/j.omtm.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao G, Wang Q, Wang L, et al. Inadvertent gene transfer of co-packaged Rep and Cap sequences during the production of AAV vector and its potential impact on vector performance. Mol Ther 2008;16:S105–S106; doi: 10.1016/S1525-0016(16)39682-4 [DOI] [Google Scholar]
- 36. Manno CS, Pierce GF, Arruda VR, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med 2006;12(3):342–347; doi: 10.1038/nm1358 [DOI] [PubMed] [Google Scholar]
- 37. Nidetz NF, McGee MC, Tse LV, et al. Adeno-associated viral vector-mediated immune responses: Understanding barriers to gene delivery. Pharmacol Ther 2020;207:107453; doi: 10.1016/j.pharmthera.2019.107453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tanaka T, Hanaoka H, Sakurai S. Optimization of the quality by design approach for gene therapy products: A case study for adeno-associated viral vectors. Eur J Pharm Biopharm 2020;155:88–102; doi: 10.1016/j.ejpb.2020.08.002 [DOI] [PubMed] [Google Scholar]