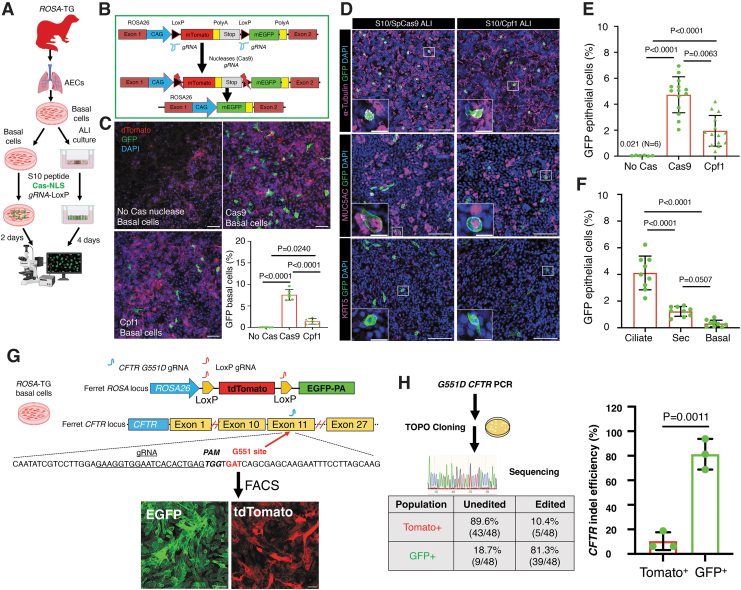
Figure 2.
S10 shuttle peptides deliver SpCas9 and Cpf1 ribonucleoproteins to ferret airway epithelial cells in vitro. (A) Schematic showing the experimental workflow for delivery of ribonucleoproteins (RNPs) to ferret basal cells and differentiated airway epithelium using the S10 shuttle peptide. Primary ROSA-TG Cre reporter ferret airway basal cells and polarized ALI cultures were delivered with the indicated Cas RNPs (SpCas9, 2.5 μM; Cpf1, 1.33 μM) containing LoxP-gRNA (2.0 μM) and S10 peptide (20 μM) for 15 min. Cultures were then washed and cultured for an additional 2 days (basal cells) or 4 days (ALI cultures) before being evaluated for genome editing by accessing EGFP+ cells. (B) Schematic illustration of the strategy for Cas nuclease RNP-mediated genome editing of LoxP sites at the ROSA-TG reporter locus in ferret airway epithelial cells. Cells were delivered with the S10 shuttle peptides complexed with Cas RNP and LoxP-gRNAs. Cas cleavage at the LoxP sites leads to the conversion of genome-edited cells from Tomato+ to EGFP+. (C) S10/Cas RNP delivery to ferret airway basal cells. Basal cells were either mock delivered with no Cas protein (top left) or delivered with SpCas9 (top right) or Cpf1 (bottom left). The genome editing efficiency was accessed as the percentage of EGFP+ cells from N = 6 donor ferrets (bottom right panel). (D) S10/Cas RNP delivery to differentiated ferret airway epithelia in ALI cultures. Representative images of immunolocalization for EGFP protein and cell-type markers for ciliated cells (α-tubulin, top panel), secretory (Sec) cells (MUC5AC, middle panel), and basal cells (KRT5, bottom panel) in epithelia delivered with S10/SpCas9 (left panel) and S10/Cpf1 (right panel) RNPs. Insets show magnifications of the boxed area in the corresponding image. (E) The efficiency of genome editing was accessed as the percentage of EGFP+ cells in ALI epithelia delivered with S10/SpCas9 and S10/Cpf1 RNPs (N = 14 ALI cultures derived from three donors). (F) The portion of genome-edited epithelial cell types in ferret ALI epithelia delivered with S10/SpCas9 (N = 9 ALI cultures derived from three donors). (G, H) Efficiency of S10-delivered CRISPR/Cas9-mediated genome editing at the CFTR-G551 locus (Gene ID: 101672484) in ferret ROSA-TG airway basal cells. ROSA-TG reporter ferret basal cells were transfected with RNP containing Cas9 protein and sgRNAs to LoxP and the CFTR-G551 locus. Following transfection, the LoxP sgRNA directs Cas9-mediated cleavage at the LoxP sites resulting in conversation of Tomato to EGFP expression. Indels at the CFTR loci were enriched by fluorescence-activated cell sorting (FACS) of EGFP+ cells (G). The CFTR indel efficiency was assessed by TOPO cloning and Sanger sequencing gave a percentage of clones carrying CFTR indel sequence(s) in Tomato+ cells (lacking LoxP-mediated conversion) and EGFP+ cells (including LoxP-mediated conversion) (H). Upper left panel: schematic workflow for the TOPO cloning assay. Lower left panel: the average indel efficiency of three donor ferrets assessed by Sanger sequencing is presented in a table. Right panel: the indel efficiency of three donors was plotted in bar graphs (N = 3). Nuclei were counterstained with DAPI in C and D. Bars equal 100 μm in C and D; 20 μm in insets of D. Data in C, E, F, and H represent the mean ± SD from three independent replicated experiments. Statistical significance was evaluated by one-way ANOVA followed by Tukey's multiple comparisons test, or two-tailed Student's t-test (H). FACS, fluorescence-activated cell sorting; RNPs, ribonucleoproteins.