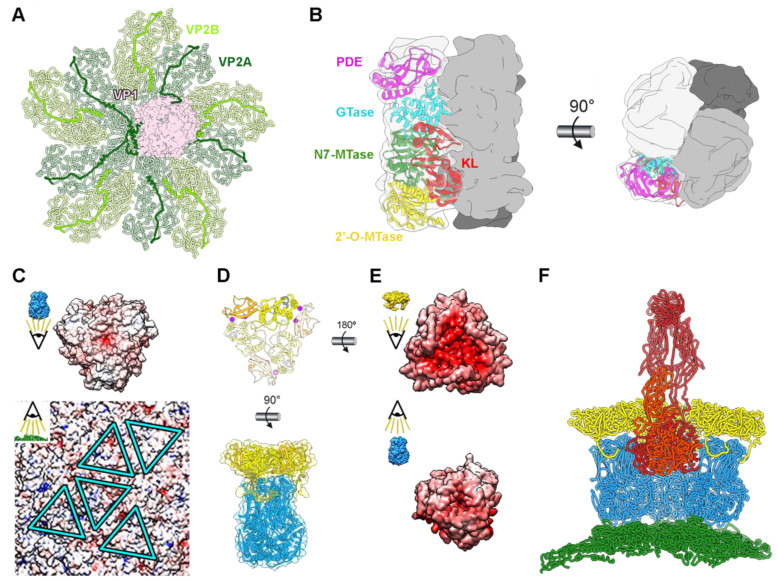
Figure 2.
Rotavirus viral protein interactions. (A) In virio interactions of VP1 RdRp (pink) and VP2 decamer (green) (PDB 6OGY). The N-termini of VP2A (dark green) and VP2B (light green) subunits are highlighted. (B) Structure of the VP3 tetramer (PDB 6O6B). One subunit is shown with transparency, with its domains represented in separate colors: kinase-like (KL) domain (red), guanine-N7 methyltransferase (N7-MTase) domain (green), 2′-O-methyltransferase (2′-O-MTase) domain (yellow), guanylyltransferase (GTase) domain (cyan), and phosphodiesterase (PDE) domain (magenta). Front (left) and top (right) views of the complex are shown [32]. (C) Electrostatic potentials of the inner surface of a VP6 trimer (top) and of the outer face of the VP2 shell (bottom). The positions for the interactions of the five quasi-equivalent trimers on the VP2 surface are marked with blue. (D) Atomic structure of the VP7 trimer (PDB 3GZT) with one subunit highlighted (top). Two calcium ions (magenta) are bound at the interface between VP7 subunits. Interaction between VP6 and VP7 trimers (bottom). (E) Electrostatic potential of the inner surface of a VP7 trimer (top) and of the outer surface of a VP6 trimer (bottom). (F) VP4 interactions. Side view of the pentamer-contacting hexamers where the spike is located. VP2 is represented in green, VP6 in blue, VP7 in yellow, and VP4 in red. Only the back VP6 and VP7 trimeric capsomers are shown.