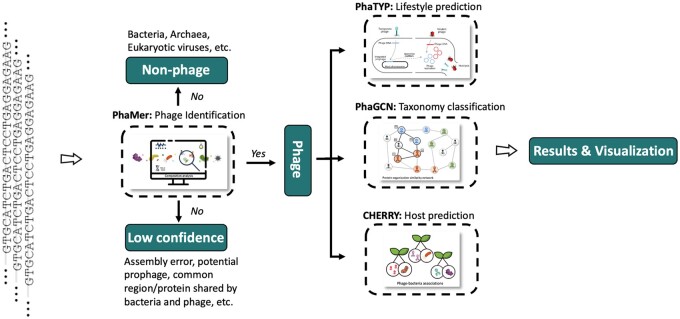
Figure 1.
The pipelines of PhaBOX. The input of PhaBOX is the FASTA file containing assembled contigs. Then, PhaMer (Shang et al. 2022) is applied for phage identification. Only the contigs predicted as phages will be used for lifestyle prediction (PhaTYP; Shang et al. 2023), taxonomic classification (PhaGCN; Shang et al. 2021), and host prediction (CHERRY; Shang and Sun 2022). Finally, we provided the predictions and visualization for users on the result page.