Chemical signaling between neurons in the brain can be divided into two major categories: fast synaptic transmission and neuromodulation. Fast synaptic transmission, mediated by amino acids such as glutamate and GABA, occurs on millisecond time scales and results in the influx of ions through ligand-gated ion channels on postsynaptic neurons (Figure 1A). Electrophysiological and optical imaging tools, including genetically encoded voltage indicators, have enabled neuroscientists to link cause (neurotransmitter release) and effect (membrane polarization) of synaptic transmission in time and space. Unlike classical neurotransmitters, neuromodulators do not produce immediate electrical effects that excite or inhibit target neurons. Instead, neuromodulators tune the intrinsic or synaptic properties of neurons, most commonly through interaction with G-protein-coupled receptors (GPCRs) (Figure 1B). Neuromodulators can escape the synaptic cleft and diffuse broadly, allowing them to influence the activity of many neurons in a state-dependent manner. Therefore, the spatial component of neuromodulator flux is fundamentally important. However, the temporal and/or spatial limitations of techniques classically used to study neuromodulation, such as microdialysis and fast-scan cyclic voltammetry (FSCV), make it difficult to interpret how neuromodulator release affects the plasticity or function of target neuronal populations on a moment-to-moment basis. Therefore, tools that can detect neuromodulators with high spatiotemporal resolution are critical for understanding their impact on neural computations that control behavior in health and disease.
Figure 1.
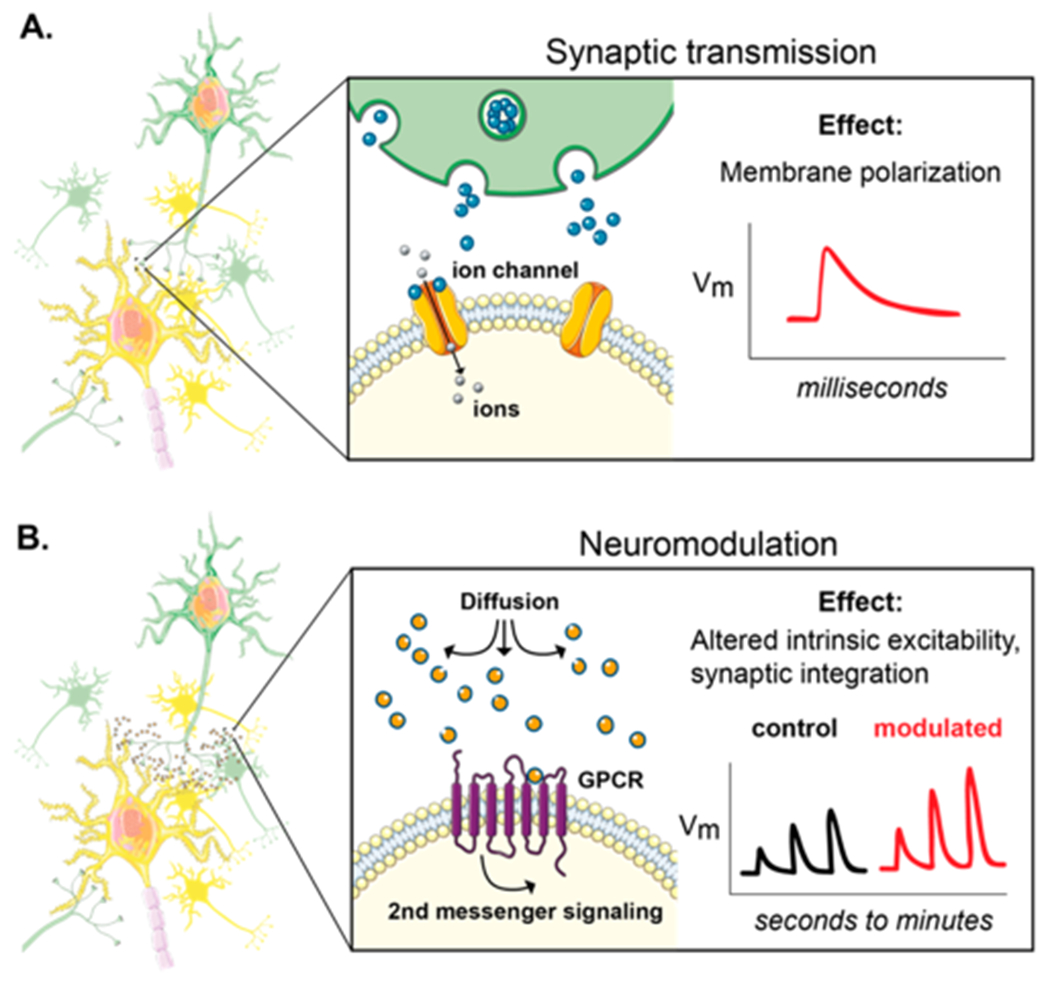
Two major modes of chemical signaling in the brain. (A) In fast synaptic transmission, neurotransmitter binds ligand-gated ion channels, allowing positively or negatively charged ions to flow into the cell. Ion flux rapidly changes the membrane potential (millisecond time scale). (B) In neuromodulation, neuromodulators can escape the synaptic cleft and diffuse, allowing them to influence broader neural networks. GPCRs are the main targets of neuromodulators and do not pass current. GPCRs engage intracellular second-messenger pathways to modulate the function of pre- or postsynaptically expressed substrates through slower mechanisms (seconds to minutes). Vm is the membrane voltage. For a more in-depth review of dopamine neuromodulation, see ref 1.
Recent advances in genetically encoded fluorescent probes have begun to address this need for rapid and localized readout of neuromodulator signaling. One approach takes a “nature knows best” strategy, in which endogenous receptor proteins are covalently conjugated to a circularly permuted fluorescent protein (cpGFP) to form chimeric receptors capable of translating ligand-binding-induced conformational changes to fluorescence signal intensity changes (Figure 2A,B). These chimeric receptors can be expressed in the plasma membrane of select neuronal populations via genetic engineering strategies. This approach (albeit using a bacterial glutamate-binding protein) was previously used to create a glutamate-sensing fluorescent probe (iGluSnFr)2 and has been expanded to the neuromodulator dopamine in two recent reports.3,4
Figure 2.
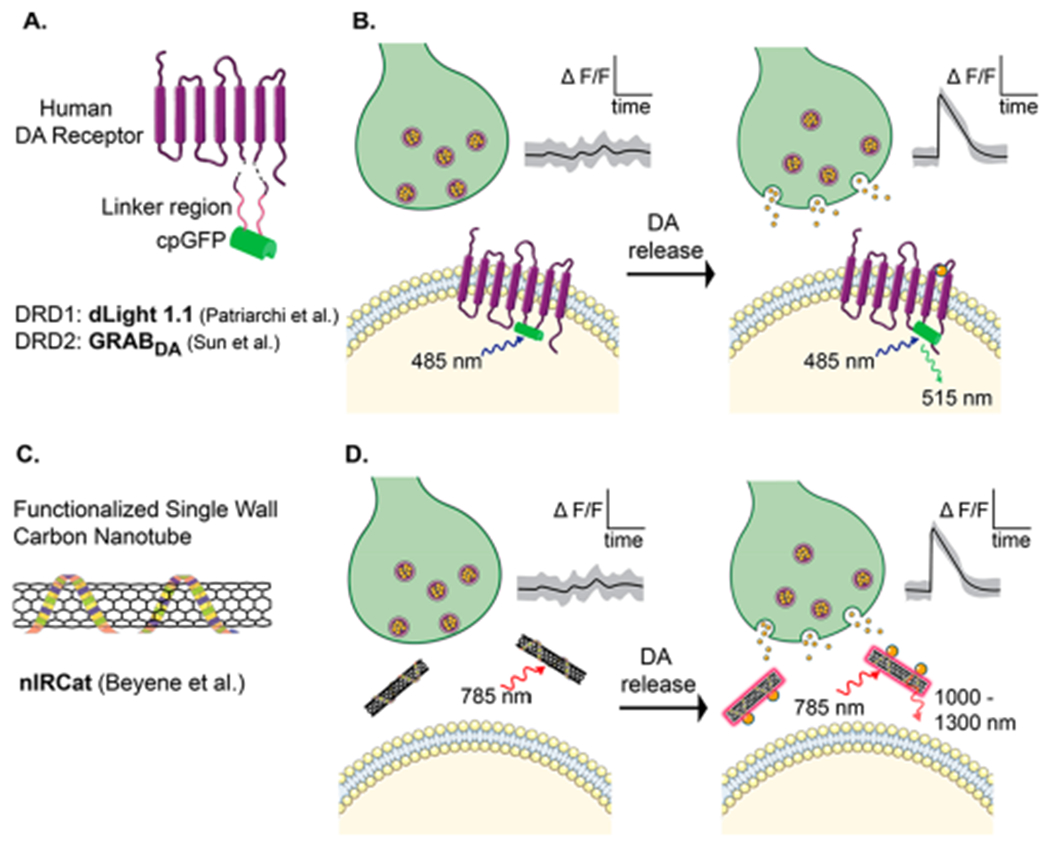
Genetically encoded fluorescent dopamine sensor design. (A) Patriarchi et al. and Sun et al. engineered human DA receptors that were covalently conjugated to a cpGFP molecule. (B) Following release by a nearby presynaptic terminal, dopamine binding induces a conformational change that increases the fluorescence of cpGFP. (C) Functionalized single-wall carbon nanotubes are used to produce an optical catecholamine sensor (nIRCat).5 (D) nIRCats localize in the extracellular space and optically respond to extracellular dopamine molecules. DA release is detected as the ΔF/F of GFP or the nIRCat signal caused by dopamine binding.
Following parallel strategies, Patriarchi et al. and Sun et al. used the human dopamine receptor (DRD) to construct fluorescent dopamine probes named dLight1 and GRABDA, respectively. Both were developed by optimized insertion of a cpGFP module into the third intracellular loop of human DRD subtypes: GRABDA variants were constructed entirely from DRD2, and the most thoroughly characterized of the dLight1 variants were constructed from DRD1. The probes demonstrate similar performance: dLight1 variants exhibit maximal turn-on fluorescence responses (ΔF/F) from 200 to 300%, whereas GRABDA reports a peak response of ~100%. dLight1 variants report a broad range of binding affinities (reported as Kd) varying from 300 nM to 1.6 μM, and GRABDA variants report a relatively narrow range of affinities (reported as EC50) of 10–130 nM. In contrast to FSCV, both show selectivity for dopamine over its molecular analogue norepinephrine, with dLight1 demonstrating ≤70-fold selectivity and GRABDA showing ≤10-fold selectivity. Both probes exhibit stable fluorescence and efficiently traffic to the plasma membrane, affording high signal-to-noise ratios that enable video-rate fluorescence imaging. Importantly, both dLight1 and GRABDA did not exhibit ligand-induced changes in cAMP, providing preliminary evidence that the probes do not interfere with endogenous GPCR signaling.
The authors demonstrated the utility of dLight1 and GRABDA across an impressive list of in vitro cell cultures, ex vivo brain slices, and awake, behaving animals (Figure 3). From a practical standpoint, the spectral profile of dLight1 and GRABDA probes fits seamlessly into existing imaging setups utilized by many neuroscience laboratories. Furthermore, the optical versatility of the probes easily enables multiplexing: for instance, in vivo dopamine sensor imaging can be combined with calcium imaging or optogenetic stimulation using red-shifted calcium indicators (jRGECO) or optogenetic actuators (ChrimsonR), respectively (see Figure 3A–C).
Figure 3.
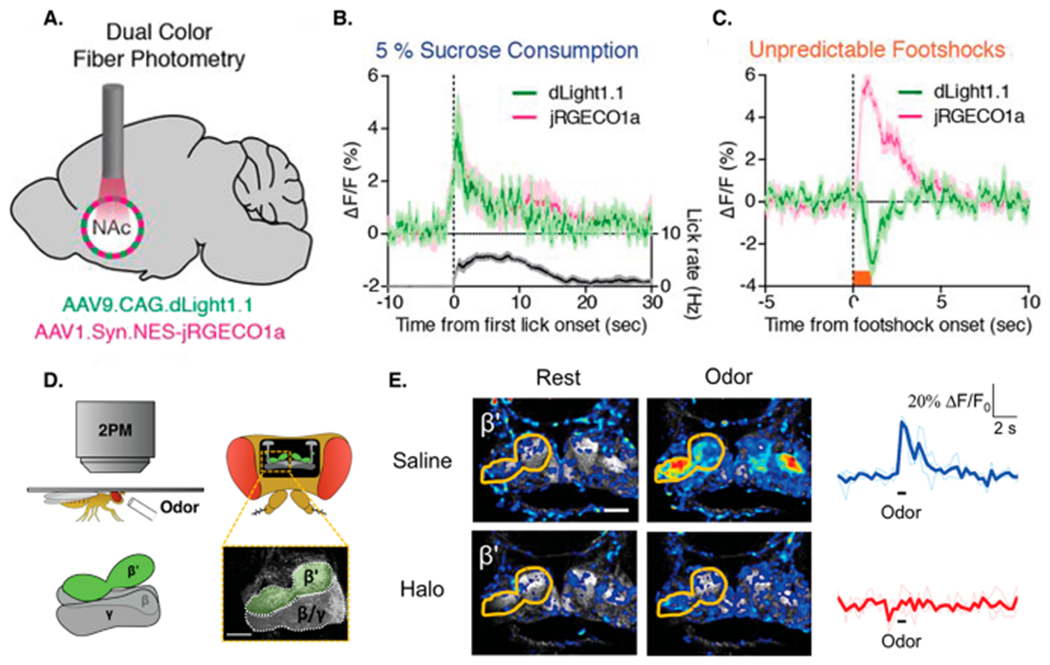
dLight1 and GRABDA in action. (A) Fiber photometry implemented in the nucleus accumbens (NAc) to measure dopamine in response to motivationally salient stimuli. dLight1.1 and red-shifted calcium indicator jRGECO1a were expressed via the delivery of AAV into mouse NAc. (B) Presentation of 50 μL of 5% sucrose elicits an increase in dopamine release, as detected by dLight1.1 fluorescence and a concordant increase in local population neural activity, reported by simultaneously recorded jRGECO1a fluorescence. (C) Unpredictable foot shocks (0.6 mA applied for 1 s) suppressed dopamine release, while the local neural activity remained high. (D) Imaging dopamine dynamics in Drosophila using two-photon microscopy. The GRABDA sensor was genetically expressed in GAL4 transgenic driver lines. (E) Presentation of odor (isoamyl acetate applied for 1 s) elicited dopamine release in the β′ lobe of the fly brain. Fluorescence responses were blocked by DR2 antagonist haloperidol (Halo), confirming that the fluorescence increase was mediated by dopamine binding. The scale bar in panel E is 25 μm. Panels A–C are reproduced with permission from ref 3. Copyright 2018 American Association for the Advancement of Science. Panels D and E are reproduced with permission from ref 4. Copyright 2018 Elsevier.
These new probes demonstrate how advances in fluorescence microscopy, genetics, and protein engineering intersect to accelerate neuroscience research. Protein-based probes rely on genetic model organisms, which motivates parallel development of synthetic approaches that do not require gene delivery and may offer more seamless deployment and orthogonal readouts to their protein-based counterparts. A recent carbon nanoparticle-based catecholamine probe, named nIRCat, provides a purely synthetic alternative to genetically encoded dopamine probes (Figure 2C,D).5 nIRCats have demonstrated optical dopamine detection in ex vivo brain slices at similar spatial and temporal resolutions achieved by dLight1 and GRABDA. Though nIRCat lacks selectivity for dopamine over norepinephrine compared to dLight1 and GRABDA, nIRCats demonstrate unaltered performance in the presence of dopamine receptor-binding drugs, thus enabling full-panel pharmacological studies of dopamine modulation. nIRCats also exhibit other advantages, including the relatively tissue-penetrating wavelength of its near-infrared emission spectra, and hours-long photostability.
To gain a holistic understanding of brain neurochemical function, scalable tools that can measure neurochemistry at spatial and temporal domains commensurate with brain function are needed. Both dLight1 and GRABDA, and new synthetic probes like nIRCat, offer complementary capabilities and have expanded the optical tool kit available to the neuroscientist in exciting ways, ushering in a new era for the study of brain neurochemistry.
ACKNOWLEDGMENTS
The authors thank Johannes de Jong for helpful discussion.
Funding
The authors acknowledge support by National Institute of Drug Abuse CEBRA Grant R21DA044010 (to L.W.), a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI) (to M.P.L.), the Simons Foundation (to M.P.L.), a Stanley Fahn PDF Junior Faculty Grant via Award PF-JFA-1760 (to M.P.L.), a Beckman Foundation Young Investigator Award (to M.P.L.), and a DARPA Young Investigator Award (to M.P.L.). M.P.L. is a Chan Zuckerberg Biohub investigator. A.G.B. and S.J.Y. are supported by a National Science Foundation Graduate Research Fellowship, and K.D. is supported by a National Institute of Mental Health postdoctoral fellowship under Grant F32MH110184.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Tritsch NX, and Sabatini BL (2012) Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron 76 (1), 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, et al. (2013) An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10 (2), 162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong WH, et al. (2018) Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360 (6396), eaat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sun F, Zeng J, Jing M, Zhou J, Feng J, Owen SF, et al. (2018) A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174 (2), 481–96e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Beyene AG, Delevich K, Del Bonis-O’Donnell JT, Piekarski DJ, Lin WC, and Thomas AW (2018) Imaging striatal dopamine release using a non-genetically encoded nearinfrared fluorescent catecholamine nanosensor. bioRxiv, DOI: 10.1101/356543. [DOI] [PMC free article] [PubMed] [Google Scholar]


