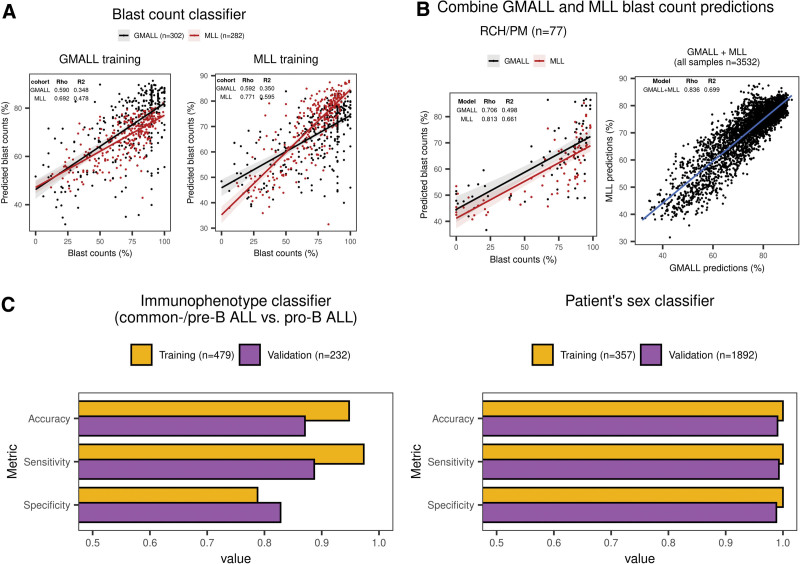
Figure 4.
ALLCatchR predicts sample blast counts, patient’s sex, and immunophenotype based on the gene expression data. (A) For GMALL (n = 302), MLL (n = 282), and RCH/PM (n = 77), sample blast counts obtained by cytology or flow cytometry were available. GMALL and MLL cohorts were separately used for training 2 classifiers in a 10-fold cross-validation scheme with the same machine learning algorithms used for subtype prediction. GMALL and MLL classifiers were validated on each other, and both were validated on the RCH/PM data. Best performing methods in terms of the RSME on the training data are shown. Training 2 classifiers on independent data sets allowed for the validation on each other and both were combined for final predictions. Blast count predictions had a good correlation to measured counts, that is, rho = 0.590 in GMALL and rho = 0.771 in MLL. Moreover, predicting MLL samples with the classifier trained on GMALL achieved a similar performance as the classifier trained on MLL samples and vice versa. (B) Because both GMALL and MLL classifiers had a good performance and were generalizable, predictions from both are combined in ALLCatchR. (C) Subclassifiers for immunophenotype and patient’s sex were developed using SVM linear and ranger machine learning models, respectively. An immunophenotype classifier was trained on GMALL samples (n = 413 common-B/pre-B and n = 66 pro-B) and validated on MLL data (n = 168 common-B/pre-B and n = 64 pro-B) with available EGIL immunophenotypes. A patient sex classifier was trained on n = 357 GMALL samples (female = 165; male = 192) analogous to the subtype classifier. For validation n = 1892 St Jude samples with known sex (female = 850; male = 1042) were used. Corresponding accuracies, sensitivities, and specificities are shown for these subclassifiers. BCP-ALL = B-cell precursor acute lymphoblastic leukemia; RSME = root mean squared error.