Fig. 1. The framework of MarsGT.
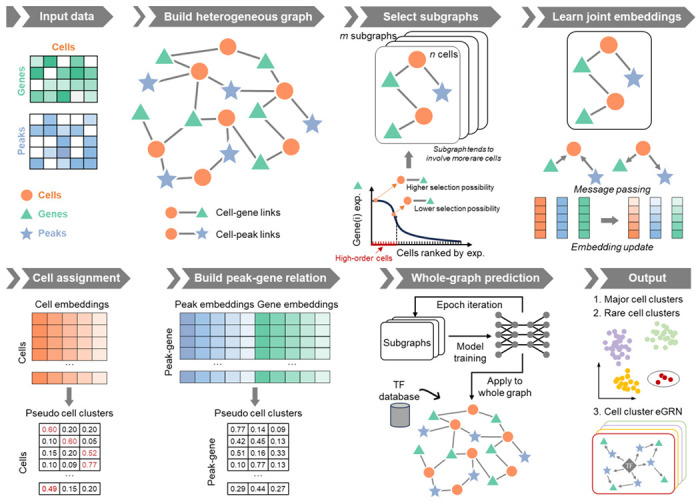
The model employs a six-step process for cell clustering and eGRN inference from matched scRNA-seq and scATAC-seq data. First, a heterogeneous graph, comprised of cells, genes, and peaks, is constructed from the original scRNA-seq and scATAC-seq data. Genes and peaks serving as edges are selected for a cell if they show high accessibility/expression within that cell and low accessibility/expression in others. The second step is learning joint embedding, which employs four graph relations to pass the message via a heterogeneous transformer. Arrows depict connections between target and source nodes. The third step involves cell assignment. Here, cell clusters are inferred from a probability matrix with rows representing cells and columns indicating pseudo-cell clusters. Cells that share the same maximum probability belong to the same cluster. The fourth step is constructing the peak-gene relationship via a matrix calculated from gene and peak embeddings, with rows denoting the regulatory potential of the peak to gene and columns indicating pseudo-cell clusters. In the final step, the trained model is applied to the entire graph. Following this, TF database information is integrated to infer cell clusters and eGRNs. Circles represent rare cell populations.
