Figure 3. Oral virologic characterization of study participants at entry.
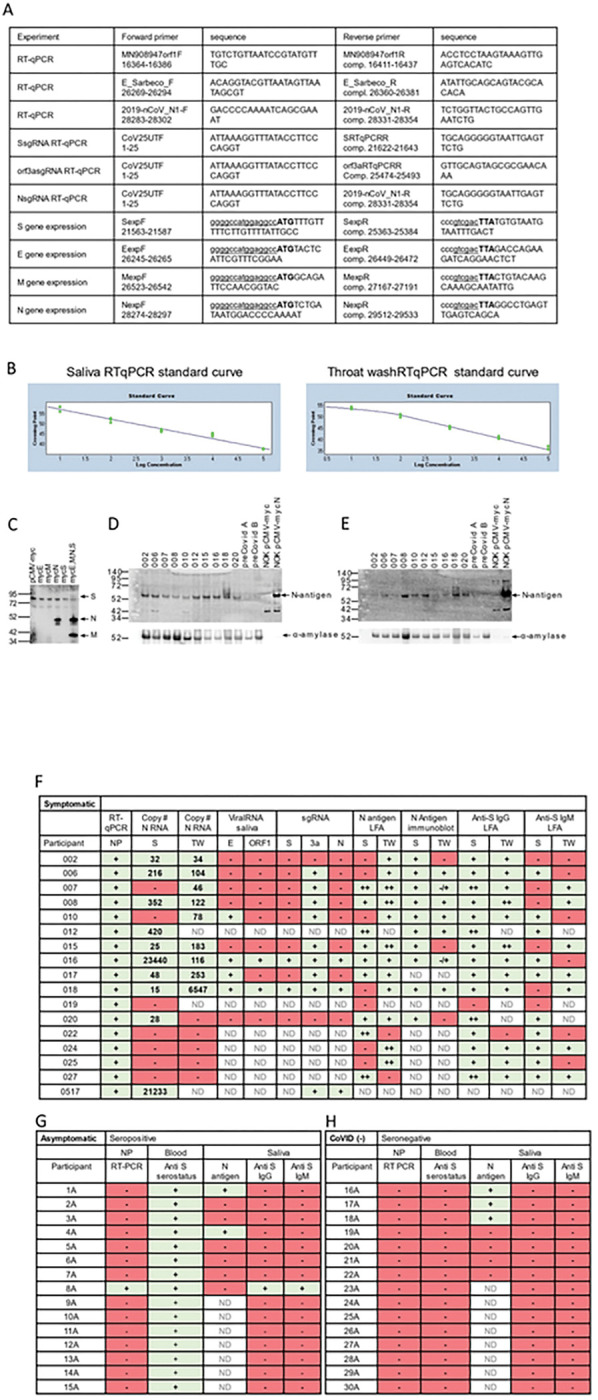
All subjects were assayed by qPCR for detection of viral RNA in the nasopharynx. Saliva/TW was assayed by qPCR for viral RNA and for LFA-based detection of N-antigen, anti–spike IgG and anti-spike IgM. A. Oligonucleotide sequences for primers used for RT-qPCR and production of cDNA for construction of expression plasmids. E_Sarbeco_F and E_Sarbeco_R have been described. The sequences of 2019-nCoV_N1-F and 2019-nCoV_N1-R, were obtained from the CDC. The SARS-CoV-2 reference sequence GenBank:MN908947 was used to determine oligonucleotide sequences and map coordinates. SfiI (ggccatggaggcc) and SalI (gtcgac) restriction enzyme sites used for cloning into the expression vector, pCMV-myc, are underlined in expression primers. The start codon and stop codons in expression primers are in bold. B. Standard curves were generated by RT-qPCR using know copy numbers of SARS-CoV-2 RNA and primers targeting the nucleocapsid (N) coding region to quantitate viral RNA isolated from patient saliva and TW. C.Immunoblot was used to detect SARS-CoV-2 structural proteins in cell culture media obtained from NOKs transfected with individual expression vectors encoding myc-tagged, SARS-CoV-2 structural proteins (S, E, M and N) or cotranfected with all myc-tagged expression vectors. Medium from cells transfected with empty vector (pCMV-myc) was used as a negative control. Proteins were detected using myc-specific antibody (mouse anti-myc, sc-40, Santa Cruz Biotechnology). D and E. Protein from subset of saliva and TW samples (respectively) was used for the detection of viral proteins. Immunoblot detection of SARS-CoV-2 N-antigen was assessed in a subset of symptomatic individuals (n=10) at baseline (top panel) using N-specific antibody (PA5–114448, LifeTechnologies, Inc). Detection of salivary alpha-amylase (anti-amylase, A8273, Sigma-Aldrich)served as a loading control (bottom panel). Saliva or TW from archived preCOVID-19 patients A and B were included as negative controls. Cell culture medium from NOK cells transfected with myc-tagged SARS-CoV-2 N-antigen expression construct was used as a positive control. Cell culture medium from NOKs transfected with empty vector was included as a negative control. N-antigen was detected at 55 KD and alpha amylase, the loading control, was detected at 58.4 KD. F.Table of symptomatic subjects (n=17) who were all nasopharynx RT-qPCR positive. RT-qPCR and LFA was also performed on saliva and TW of these subjects. Viral RNA copy number (molecules of virus RNA) in 25 microliters of saliva or TW was determined using N-specific primers. Green highlight indicates a positive result. Red highlight indicates a negative result. ND indicates not determined. G. Asymptomatic participants (n=15) were all anti spike IgG positive in their blood. H. COVID-19 negative participants (n=15) were anti-spike negative and nasopharynx negative.
