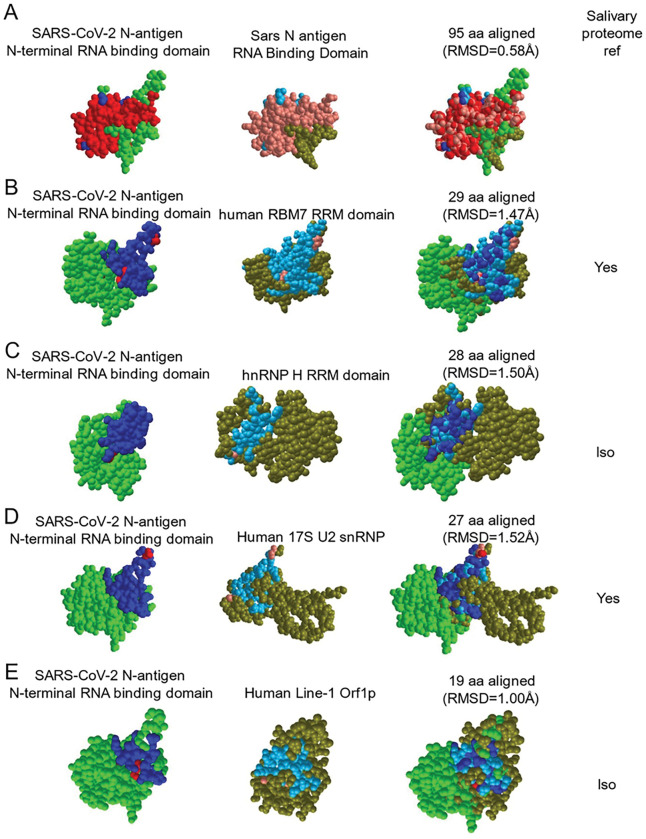
Figure 4. The SARS-CoV-2 nucleocapsid mimics host RNA binding proteins that are expressed within the salivary proteome and may be responsible for cross reactivity in LFA assays.
VAST+ was used to generate RNA binding protein structures demonstrating 3D similarity to the SARS-CoV-2 N-antigen. Simultaneous alignment generated molecular protein pairs between N-antigen (first column) and host RNA binding proteins (second column). Non-aligned N-antigen amino acids are rendered in green, conserved aligned amino acids are rendered in red and aligned, non-conserved amino acids are rendered in blue. Non-aligned host protein amino acids proteins are rendered in olive green, conserved aligned amino acids are rendered in pink and aligned, non-conserved amino acids are rendered in light blue. The spatial arrangement of overlapping amino acids can be seen in the merged sequence alignment (third column). The number of structurally aligned amino acids (conserved and non-conserved) is shown. The root mean square deviation (RMSD) is used as a measurement between atoms in the backbone of the molecular structures and is listed in angstroms above each merged image and detection of the host protein within the salivary proteome is displayed as yes in the fourth column while detection of a related isoform within the salivary proteome is displayed as ISO.