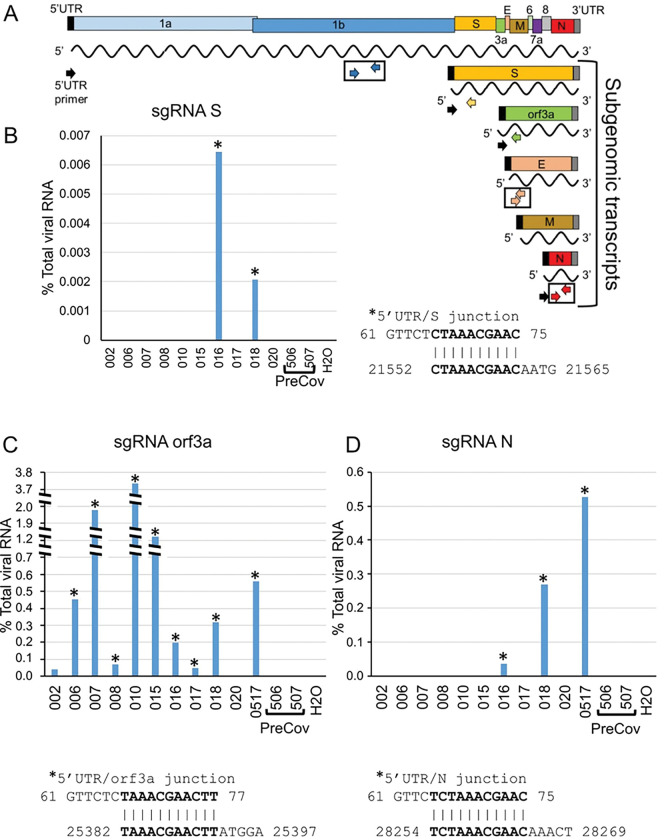
Figure 5. SARS-CoV-2 subgenomic RNAs (sgRNA) are detected the saliva of symptomatic COCID patients.
A. Schematic diagram showing the RNA genome of SARS-CoV-2 (~30 kb) and the subgenomic RNAs encoding major viral structural proteins (S, E, M and N) as well as orf3a. Detection of SARS-CoV-2 RNA by RT-qPCR was performed using primer pairs corresponding to orf1 (blue arrows), E (peach arrows) and N (red arrows) coding regions. SgRNA was detected using a forward primer corresponding to the 5’UTR (black arrow) and either S-specific (yellow), orf3a-specific (green) or N-specific (red arrow) reverse primers. The RT-qPCR signal generated with N1 primers (red arrows) were used to calibrate levels of sgRNA. B, C and D. The percent levels of S, orf3a or N sgRNA determined using the 5’UTR primer and the specific sgRNA reverse primer are shown. Percent sgRNA levels relative to total viral RNA levels are determined as follows: 2^-(sgRNA Ct/N Ct) X 100. Direct sequencing of RT-qPCR amplified products was performed to determine the junction sequence of the 5’UTR and the coding region of each sgRNA. Sequencing information obtained from samples is indicated with an asterisk. The overlap sequence between the 5’UTR and each specific sgRNA is indicated.