Figure 3.
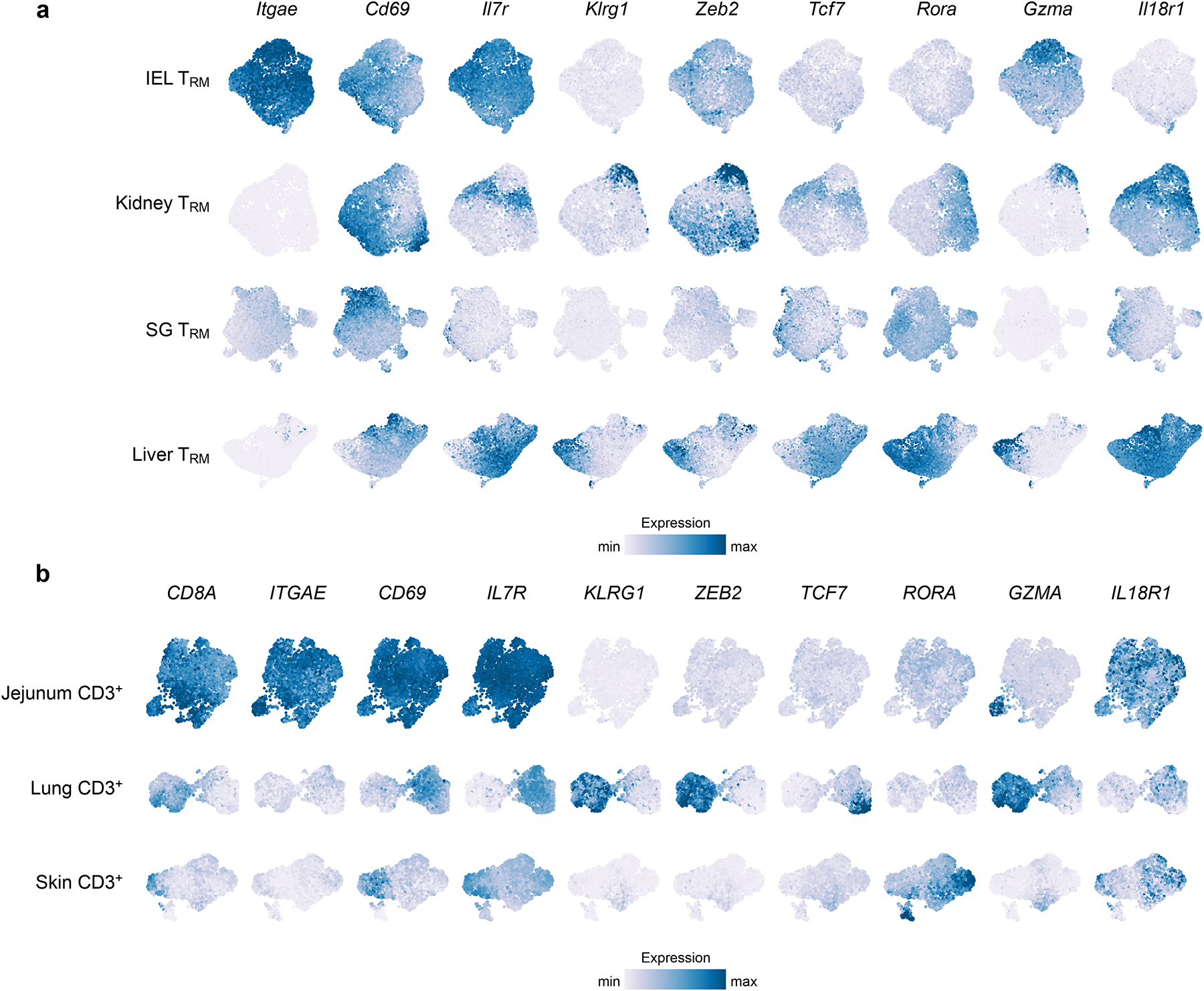
scRNA-seq reveals intertissue and intratissue heterogeneity. TRM cells within a single tissue display varying expression of genes associated with effector and memory potential, showing that distinct subsets of TRM cells exist. Scales for each gene are consistent across tissues to allow for comparison within and among tissues. Data from (a) Crowl et al. 2022 (scRNA-seq of murine IV− CD8+ P14 T cells from IEL, kidney, SG, and liver at day 32 post infection with LCMV Armstrong) and (b) Poon et al., 2023 (scRNA-seq of human CD3+ T cells of two donors). Donor D551: Jejunum and Lung; Donor D492: Skin.
For visualizing the intra-tissue heterogeneity, each tissue dataset (downloaded from GEO GSE182275 and GSE206507) was normalized separately using sctransform with method = “glmGamPoi” in Seurat102. Dimensional reduction was performed using the RunPCA function in Seurat following by RunUMAP using the first 20 PCAs. In addition, data imputation was performed using MAGIC103 using the default settings and the exact solver. Manually selected genes to highlight inter- and intratissue heterogeneity were plotted. Color scale was limited to the 98th expression percentile.
