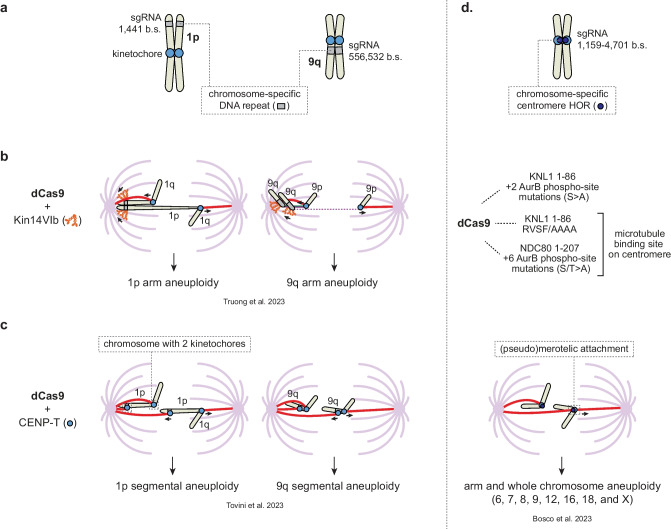
Fig. 4.
dCas9-based methods for mis-segregating specific human chromosomes. a–c dCas9 fused to either P. patens Kinesin14VIb (Kin14VIb) (b), or human CENP-T1-243 (c) has been used in combination with a sgRNA targeting chromosome-specific DNA repeats in chr1 and 9 for inducing chromosome-specific mis-segregation and aneuploidy. d A third strategy targets chromosome-specific higher order repeats (HOR) of centromeres with unique sgRNAs to recruit dCas9 fused to either KNL11-86/S24A;S60, KNL11-86/RVSF/AAAA, or NDC801-207/6xS/T>A. We propose that dCas9-KNL1-86/RVSF/AAAA and dCas9-NDC801-207/6xST>A together with a centromere-specific sgRNA create an ectopic microtubule binding site on the centromere, which can lead to (pseudo-) merotelic attachment of the targeted chromosome during mitosis. Whole chromosome and chromosomal arm aneuploidies for chr6, 7, 8, 9, 12, 16, 18, and X could be generated using this approach. Number of predicted sgRNA binding sites (b.s.) based on the T2T genome assembly are indicated in a, d. AurB, Aurora B