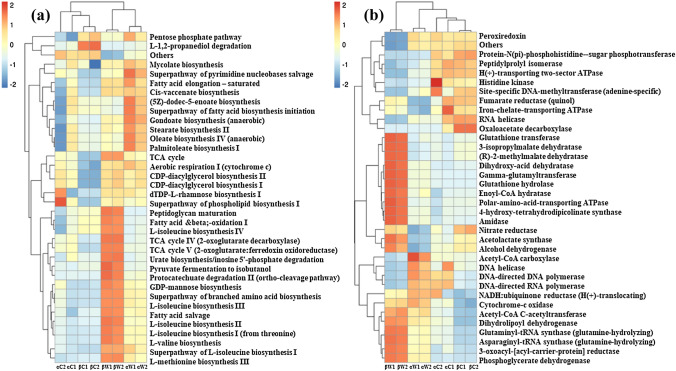
Fig. 6.
Abundances of UniRef90 families and predicted metabolic pathways were represented using PICRUSt2 (Douglas et al. 2020). a Functional pathway prediction, b predicted enzymes of microbial communities in microbiome samples. A total of 422 pathways were enriched, with 35 complete pathways depicted in the heatmap. Gut samples from site-α exhibited pathways for L-isoleucine, cis-vaccenate, stearate, and menaquinone and stress protectant ectoine biosynthetic pathway. In the water samples from both sites, enriched pathways included L-isoleucine biosynthesis, L-isoleucine biosynthesis, L-valine biosynthesis involved in immunological function; The fish gut samples from both sites displayed the presence of cellulose, lysophospholipase, lysozyme, and chitinase enzymes, involved in the digestion of organic matter. The samples from site-β exhibited a higher predominance of nitrate reductase. In the water sample from site-α, the presence of lignin-degrading enzymes, such as peroxiredoxin and glutathione peroxidase, indicated the prevalence of debris and organic matter at the polluted site