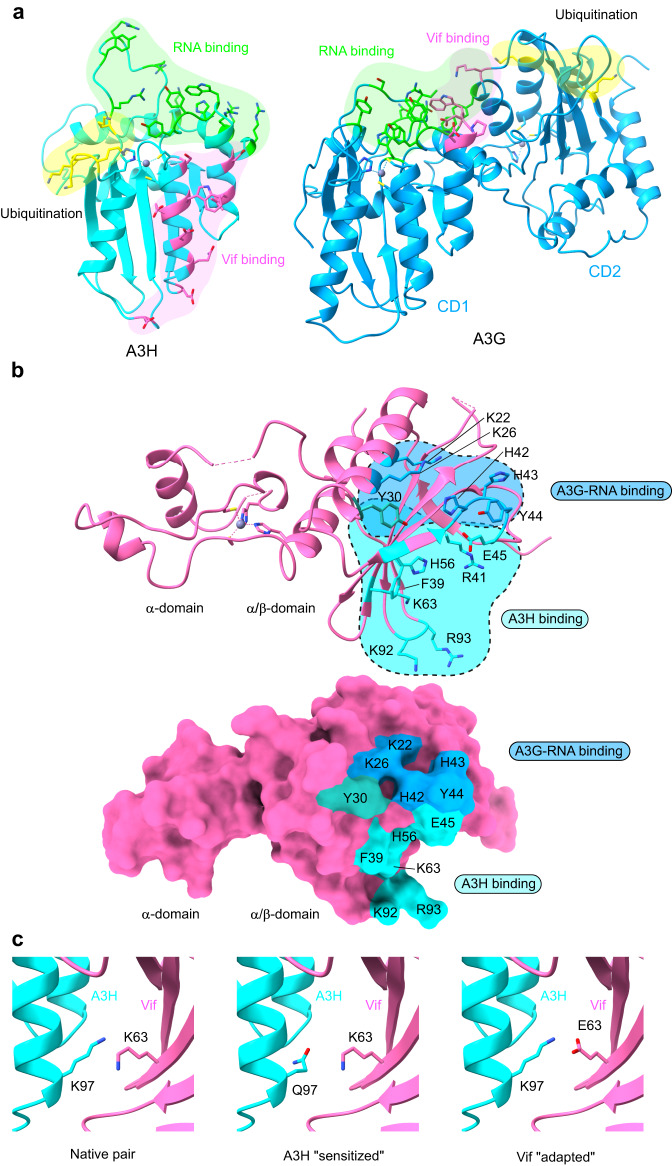
Fig. 5. Comparison of A3H-Vif and A3G-Vif complex and insights into determinants that modulate A3H-Vif interaction.
a Comparison of surface areas responsible for Vif binding, RNA binding, and ubiquitination in A3H (left) and A3G (right). b Vif surface residues responsible for A3H binding (sticks in cyan) and A3G-RNA binding (sticks in blue). Y30 is the only residue involved in both complexes (stick in pink). c Atomic model showing key amino-acid contacts modulating the A3H-Vif interaction. In native human A3H and HIV-1 Vif pair, A3H K97 is in close proximity to Vif K63, resulting in unfavorable contact (left). In sensitized A3H with K97Q mutation, the glutamine forms a hydrogen bond with Vif K63, thus stabilizing the interaction (middle). Alternatively, in a gain-of-function HIV strain that has adapted to A3H-mediated HIV restriction, Vif E63 can form an electrostatic interaction with K97, thus stabilizing the interaction (right).