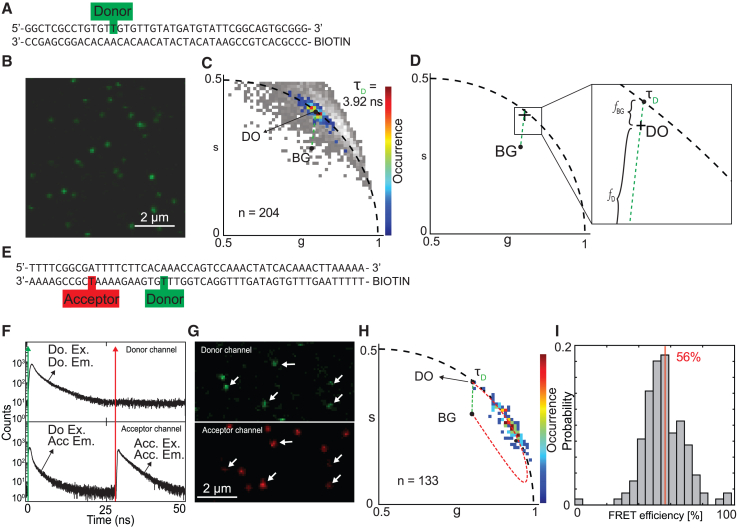
Figure 2.
Determination of FRET-labeled dsDNA FRET efficiency. (A) dsDNA strand with a single donor label on the sense strand. (B) Microscopy image using 485-nm excitation revealing single particles. (C) Pixel-binned particle phasor analysis on the donor-labeled dsDNA reveals its photon-weighted center of mass () revealing the donor lifetime of 3.92 ns ( upon extension of the fraction line to reveal its pure contributing species. Pixel-based phasor with a 10-photon/pixel threshold shown in gray. (D) Illustration of the contributing species to the phasor () location ( and ) of the donor-only-labeled dsDNA. (E) dsDNA with on the antisense strand both donor and acceptor present and a biotin tag on the 5′ end used for immobilization on the glass. (F) Pulsed interleaved excitation (PIE) measurement of FRET-labeled dsDNA used for colocalization. (G) Intensity image of double-labeled dsDNA showing a single region of interest for both donor and acceptor channel as determined by colocalization of both donor and acceptor channels for donor and acceptor excitation, respectively. White arrows indicate colocalized particles. (H) Phasor plot of imaged FRET-labeled dsDNA strands showing the autofluorescence phasor (), the donor-only phasor , and the quenching trajectory (red dashes) connecting the phasor with the phasor (see Eqs. 22 and 23). The fraction line between en (green dashes). (I) FRET efficiency histogram derived from the FRET trajectory. Vertical red line visualizes the average value.