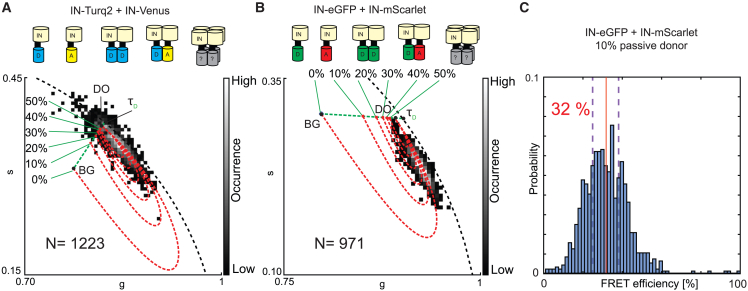
Figure 4.
FRET trajectory analysis of HIV-1 particles containing donor-labeled IN and acceptor-labeled IN using particle-based pixel binning. (A) Phasor analysis of viral HIV-1 particles containing both donor- and acceptor-labeled IN (IN-Turquoise2+IN-mVenus) forming an unknown mix of multimers including monomers and dimers and potentially higher-order multimers. The donor-only (DO) phasor (g = 0.82, s= 0.37 and background (BG) phasor (g = 0.78, s= 0.31) determine the starting point and end of the FRET trajectory. Variations of the donor-only signal contribution in the analyzed particles result in a variety of FRET trajectories and FRET trajectory end points (red dotted line). (B) Phasor analysis of viral HIV-1 particles containing both donor- and acceptor-labeled IN (IN-eGFP+IN-mScarlet) forming an unknown mix of multimers including monomers and dimers and potentially higher-order multimers. The DO (g = 0.90, s = 0.29) and BG (g = 0.79, s = 0.31) determine the starting point and end of the FRET trajectory. Variations of the donor-only signal contribution in the analyzed particles result in a variety of FRET trajectories and FRET trajectory end points (red dotted line). In both (A) and (B) the pure monoexponential lifetime of the donor () and BG determine by intensity fraction the position of DO on their connecting fraction line (green dotted line). (C) FRET histogram for (B) extracted from the 0, 10, and 20% passive donor contribution FRET trajectories of which the 0 and 20% histogram averages are indicated by purple dashed lines and the 10% passive donor histogram (shown) average is depicted as a vertical full red line with its value.