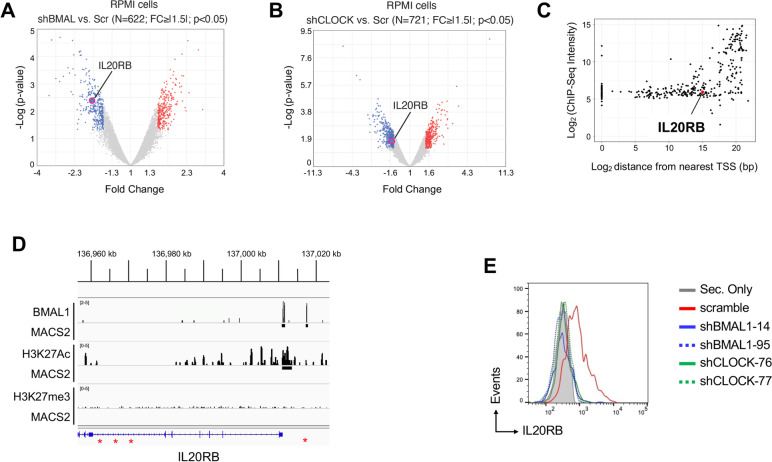
Fig. 4.
Interleukin 20 receptor, beta subunit (IL20RB) is a direct target gene of the circadian clock machinery in T-ALL. A-B Volcano plot of differentially expressed genes in shBMAL1 or shCLOCK (D) vs. shScramble T-ALL cells. Red (up-)/Blue(down-regulated) dots represent statistically significant genes (q-value < 0.05). N, number of significantly differentially expressed genes. FC, fold change (cutoff, |1.5|). C Location of predicted BMAL1 sites relative to transcription start sites (TSS) and ChIP-seq signal intensity (ChIP-seq score in Log2). In the plot, each dot represents BMAL1 peaks within 1 Kb around the TSS. The ChIP-seq signal over IL20RB gene in the RPMI-8402 cells is highlighted in red. D ChIPseq tracks for BMAL1, H3K27me3 and H3K27Ac over the human IL20RB 5’ region in the RPMI-8402 cells. Peaks of aligned reads over the IL20RB locus are shown along with MACS2 peak calls (p-value ≤ 0.05). The active genomic regions are identified by the H3K27Ac histone mark. The canonical “E-box” binding site for CLOCK-BMAL1 in the IL20RB locus are highlighted in red. E Protein expression level of Interleukin 20 receptor (IL20R) by flow cytometric analysis in the SUP-T1 cells transduced with two different clones of shRNA/GFP lentiviral constructs against BMAL1 or CLOCK genes or scramble control as indicated and in vitro cultured for 7 days. GFP + alive cells were measured after seven days from the transduction and identified for DAPI exclusion by flow cytometry