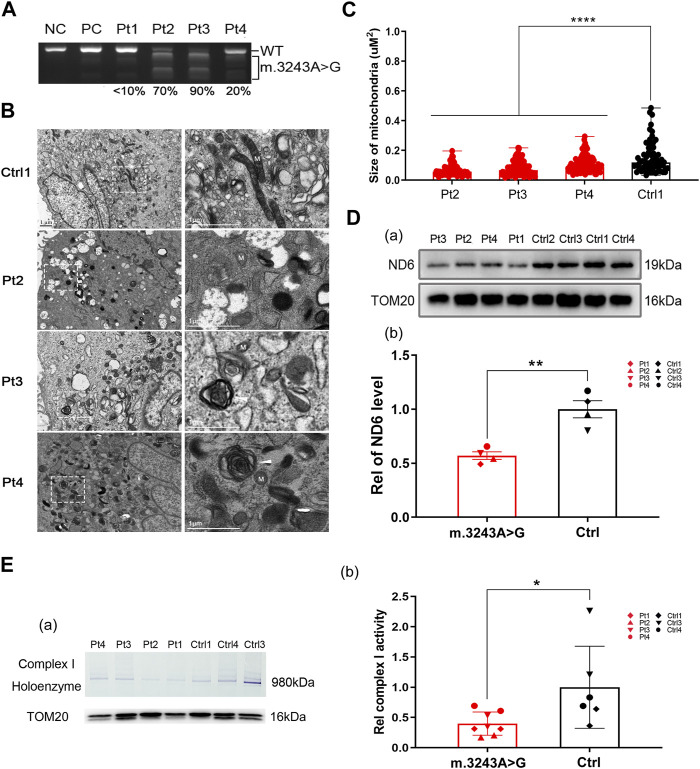
FIGURE 1.
The decreased expression of ND6 may account for the deficiency in complex I observed in the m.3243A>G fibroblasts. (A) The m.3243A>G mutation rate of m.3243A>G fibroblasts was determined by RFLP analysis compared to negative control and positive control. (B) Mitochondria (M) in m.3243A>G and control fibroblasts were visualized by transmission electron microscopy. Arrowheads indicate myelin figures. (C) The size of the mitochondria in m.3243A>G and control fibroblasts was measured as described in the materials and methods section (number of mitochondria counted: m.3243A>G n = 74–109, Control n = 63; data were analyzed using a one-way ANOVA with Bonferroni post hoc test; error bars represent mean ± SEM; ****p < 0.0001). (D) Western blotting (A) and quantification (B) for relative expression level of ND6 protein in m.3243A>G and control fibroblasts (m.3243A>G n = 4, Control n = 4; data were analyzed using a Student’s t-test; error bars represent mean ± SEM; **p < 0.01). (E) BN PAGE (In-gel activity) analysis (A) and quantification (B) of the complex I activity in m.3243A>G and control fibroblasts using SDS-PAGE analysis of TOM20 protein as mitochondrial loading marker (m.3243A>G n = 4, Control n = 3; data were analyzed using a Mann Whitney U test; error bars represent mean ± SEM; *p < 0.05). ANOVA, analysis of variance; BN-PAGE, blue native polyacrylamide gel electrophoresis; RFLP, Restriction fragment length polymorphism; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SEM, standard error of the mean.