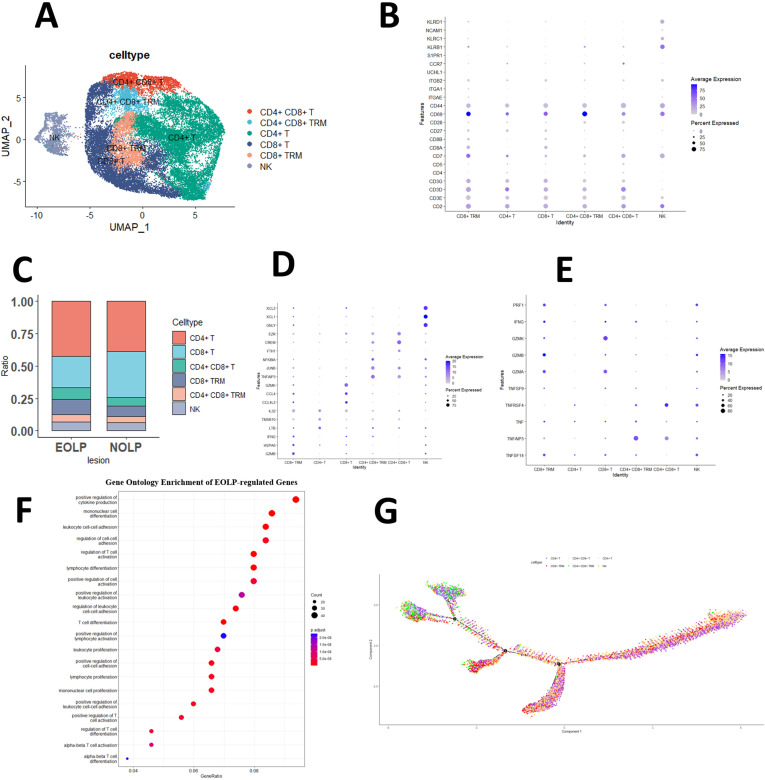
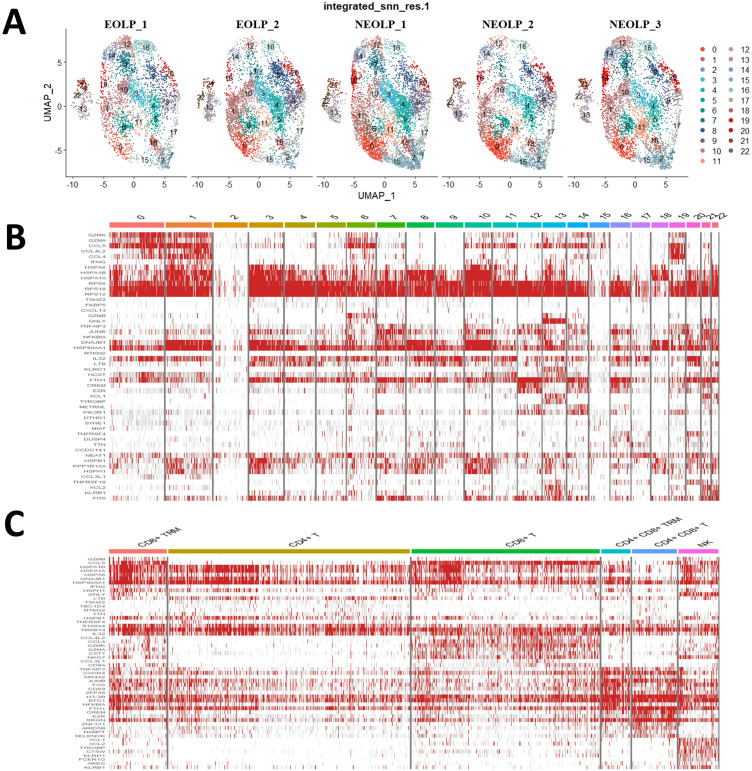
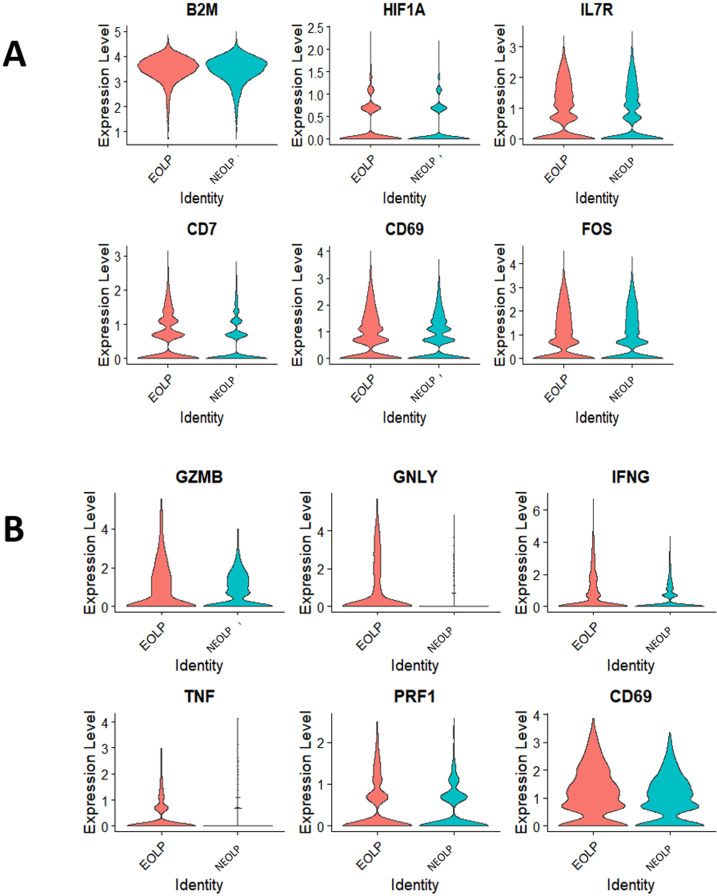
Figure 2. CD8+ Trm in oral lichen planus (OLP) patients have different transcriptomic landscapes in different clinical presentations.
(A) Uniform manifold approximation and projection (UMAP) plot of NK/T cells colored by subtype. (B) Dot plot of marker genes in NK/T cells subsets. (C) Proportion of NK/T cell subsets in OLP with different clinical manifestations. (D) Dot plot of significant differentially expressed genes (DEGs) in NK/T cells subsets. (E) Dot plot of differential expression of inflammatory cytokine product genes in NK/T cell subsets. (F) Dot plot of GO enrichment analysis of DEGs between erosive oral lichen planus (EOLP) and non-erosive oral lichen planus (NEOLP). (G) Pseudotime trajectory of NA/T cells in OLP.