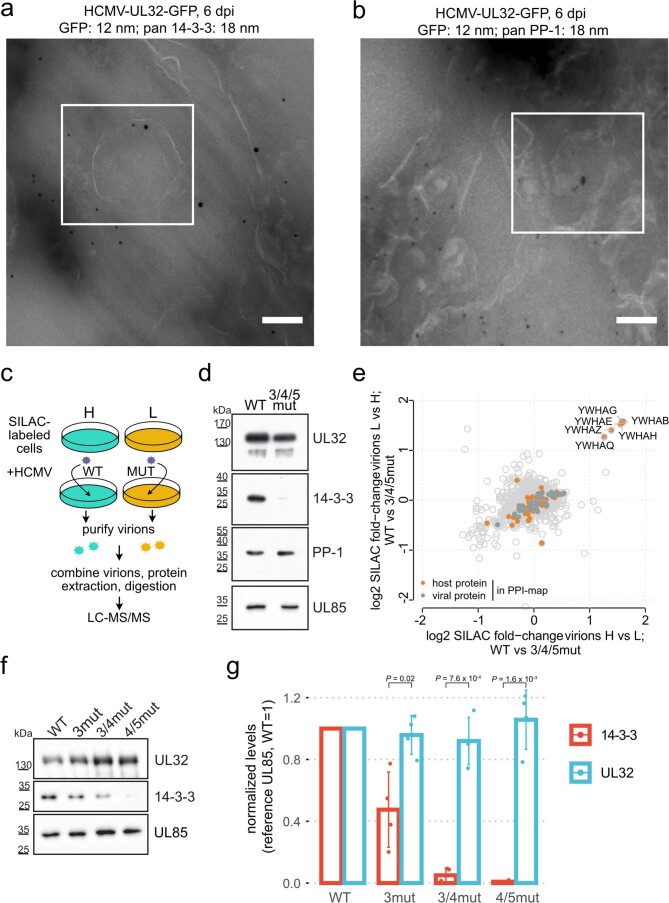
Extended Data Fig. 5. Characterization of the 14-3-3 binding-deficient viral mutants.
(a, b) EM images of the intracellular environment from HCMV-UL32-GFP infected HELFs (MOI=5) prepared using Tokoyasu method. Ultrathin sections were stained with immunogold against GFP (12 nm gold) and pan 14-3-3 or PP-1 (18 nm gold), as indicated. Scale bars: 100 nM. Magnified views of the boxed regions in white are depicted in Fig. 4b. (c) Experimental workflow for SILAC-based comparison of virion protein content between mutant and WT viruses. The experiments were performed in label-swap duplicates. (d) Purified virions from WT or mutant viruses harbouring alanine substitutions in the ¾/5 region (see also Fig. 4c) were assessed for UL32, PP-1, 14-3-3 and UL85 levels by immunoblotting. Control experiment to panel (e). (e) SILAC-based proteomic comparison of log2 protein fold-changes comparing purified particles of WT and 14-3-3 binding mutants for both replicates, based on n=2 biological replicates. (f, g) Western blot analysis of abundance levels of selected proteins of purified virions for different 14-3-3 binding site mutant viruses. Exemplary blots in (f) and quantification based on n=2 biological replicates (with n=2 technical replicates) in (g). The height of the bars corresponds to the mean and the error bars to the standard deviation. P-values based on two-sided t-test without multiple hypothesis correction. Sites 4/5 are most important for 14-3-3 incorporation.